Overview
QPDP is a 32-qubit quantum computing platform that revolutionizes personalized medicine by integrating genomic profiling, pharmacokinetic modeling, drug-drug interaction analysis, and adverse drug reaction prediction into a unified quantum framework.
View Complete Technical Report →
Patient Profile Generation
QPDP generates comprehensive patient profiles integrating genomic data, demographics, comorbidities, current medications, and allergy profiles.
Profile Components
| Component | Data Points | Purpose |
|---|---|---|
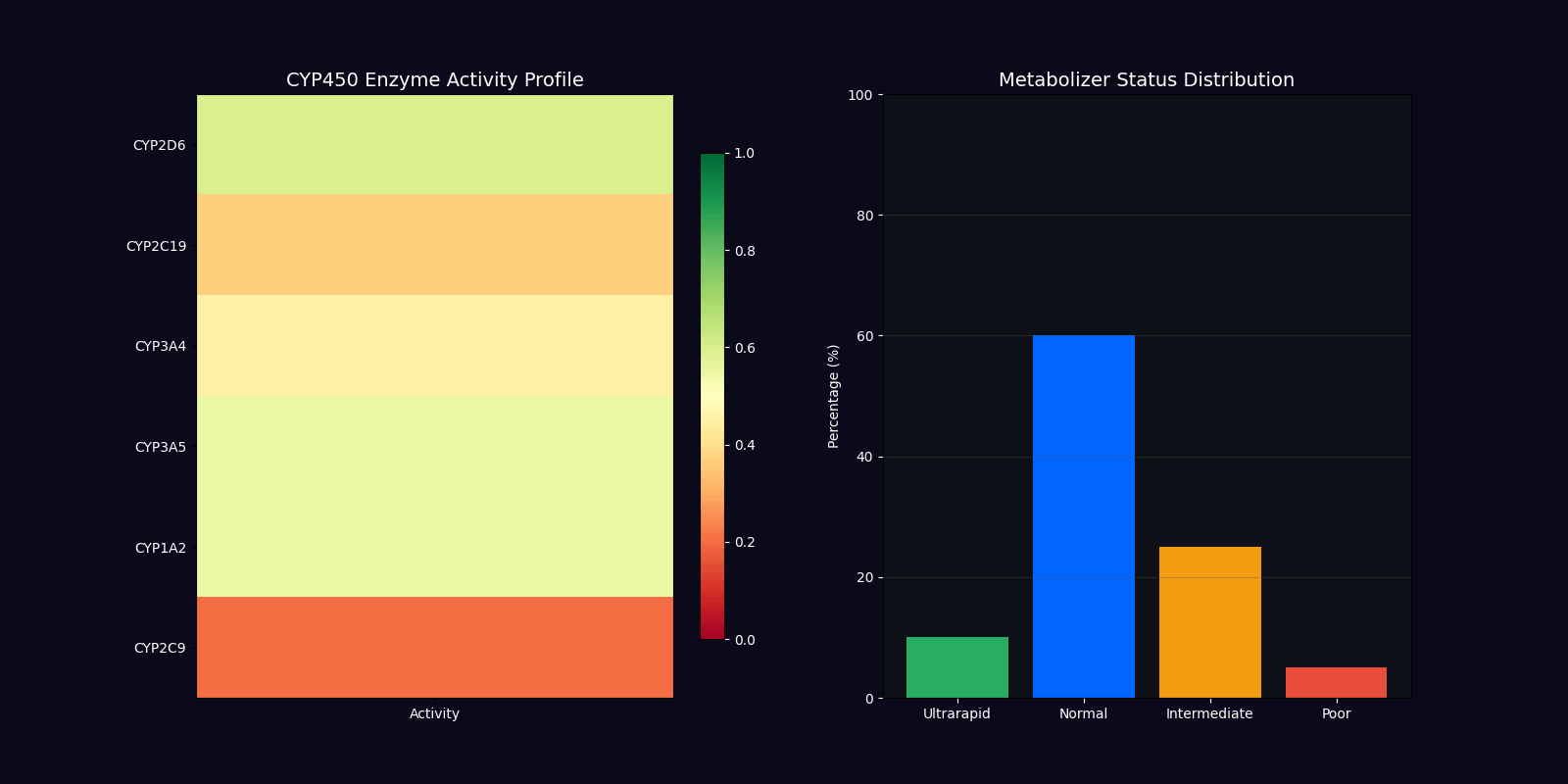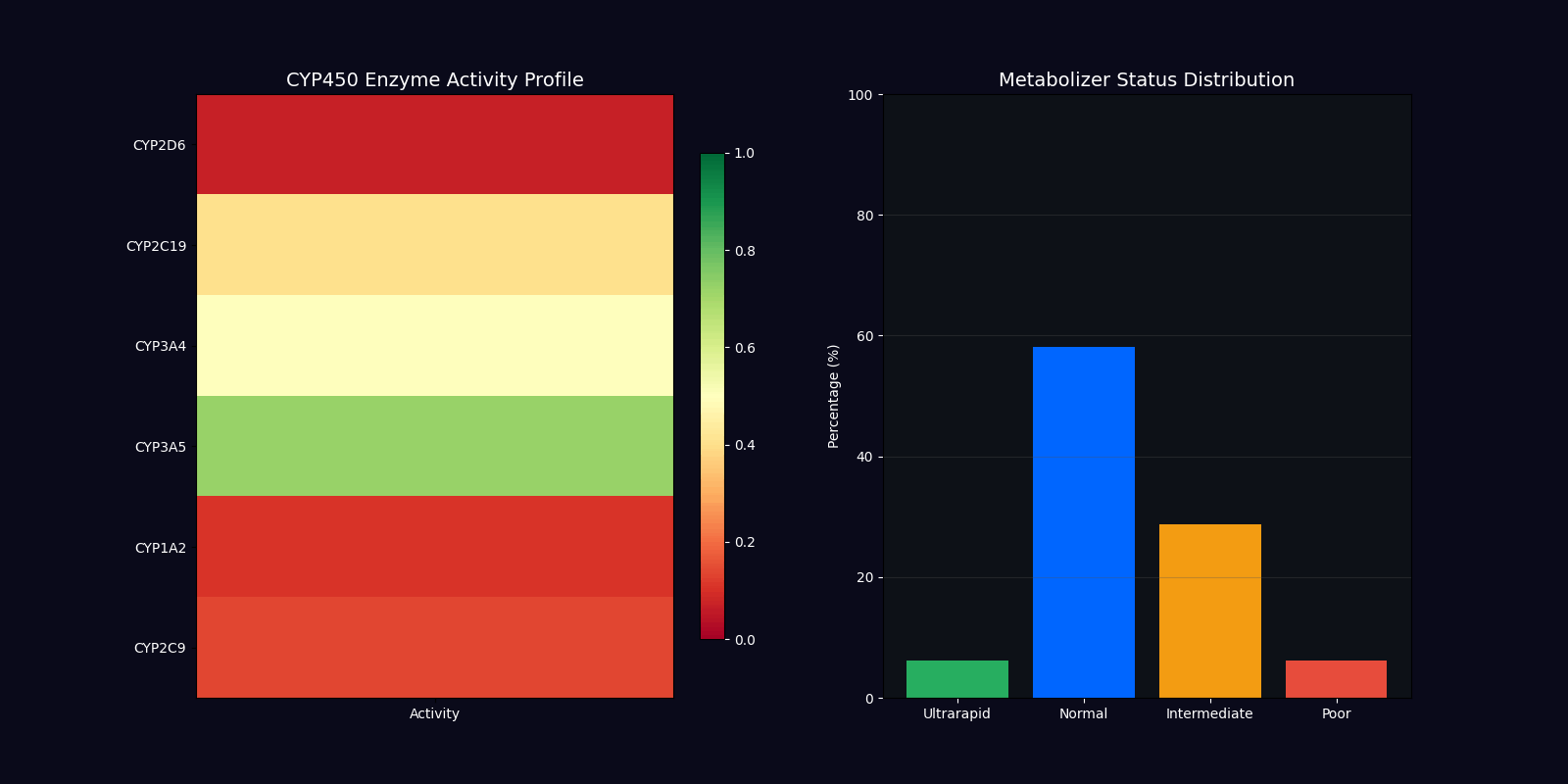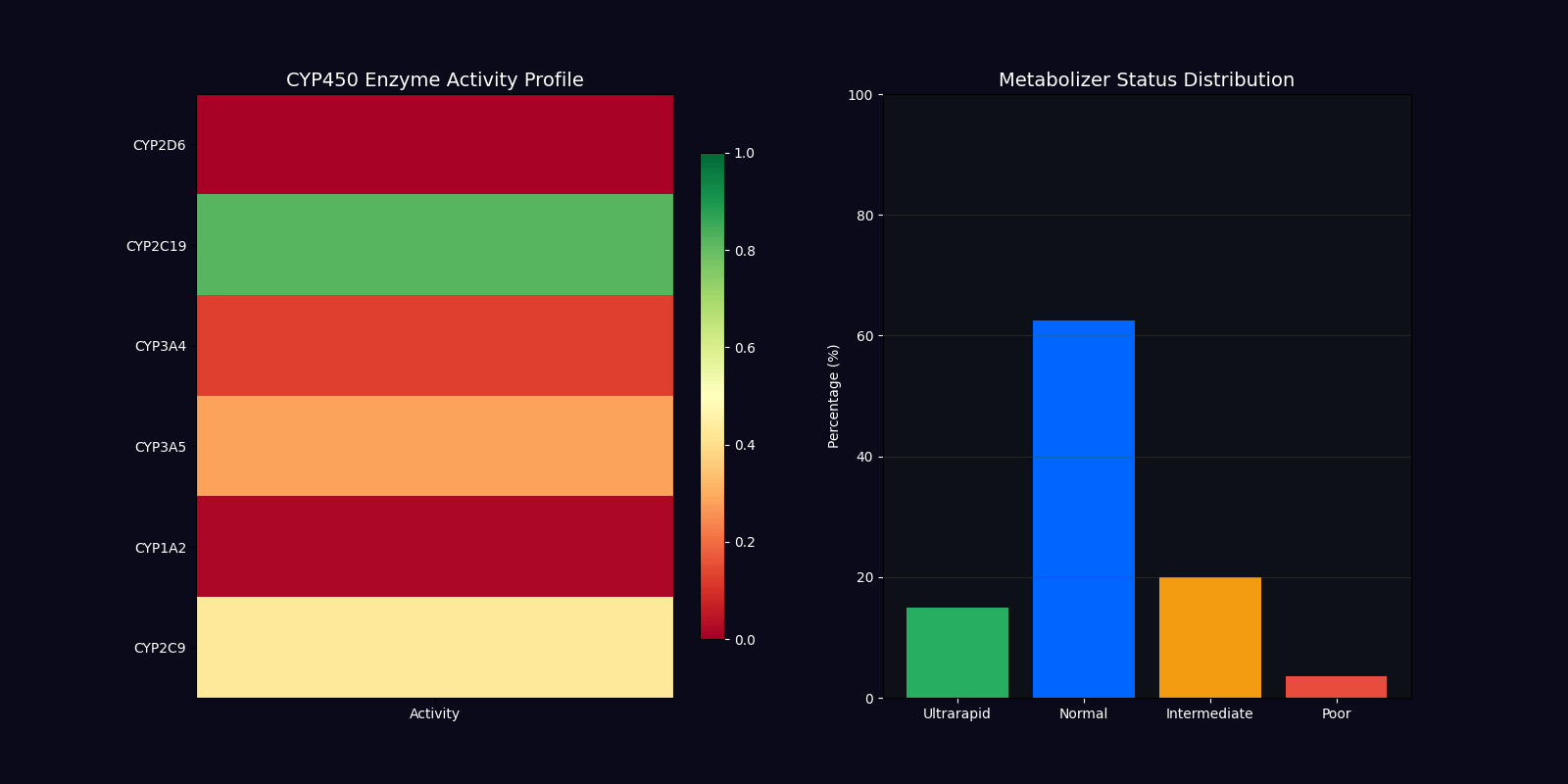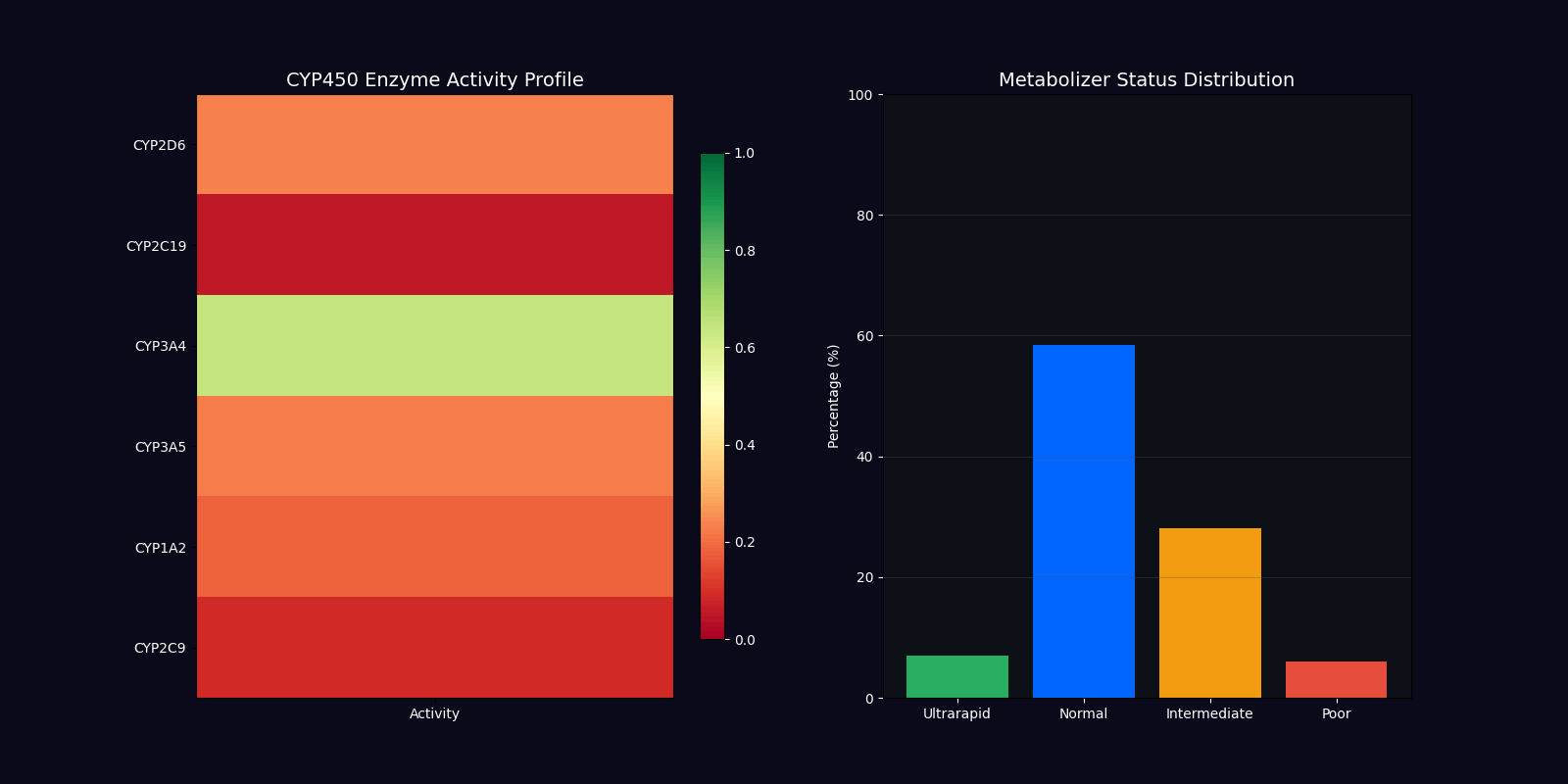
| CYP450 Genotyping | 2D6, 2C19, 3A4, 3A5 | Metabolizer status determination |
| Demographics | Age, weight, BMI | Dosing calculations |
| Comorbidities | Cardiovascular, renal, hepatic, metabolic | Risk adjustment |
| Current Medications | Active prescriptions | Interaction detection |
| Allergy Profile | Known drug allergies | Safety screening |
Genomic Analysis
Advanced pharmacogenomic analysis using CYP450 enzyme variants to predict drug metabolism and response.
Metabolizer Status Classification
Ultrarapid Metabolizers
Increased enzyme activity requiring higher doses for therapeutic effect. Dose factor: 1.5x standard.
Normal Metabolizers
Standard enzyme activity with typical drug response. Dose factor: 1.0x standard.
Intermediate Metabolizers
Reduced enzyme activity requiring moderate dose adjustments. Dose factor: 0.7x standard.
Poor Metabolizers
Minimal enzyme activity requiring significant dose reduction. Dose factor: 0.4x standard.
CYP450 Enzyme Variants
| Enzyme | Substrates | Clinical Impact |
|---|---|---|
| CYP2D6 | Codeine, Tramadol, Metoprolol, Fluoxetine | Pain management, cardiovascular, psychiatry |
| CYP2C19 | Clopidogrel, Omeprazole, Diazepam | Antiplatelet therapy, GI protection |
| CYP3A4 | Statins, Calcium channel blockers, Immunosuppressants | Cardiovascular, transplant medicine |
| CYP3A5 | Tacrolimus, Midazolam | Immunosuppression, anesthesia |
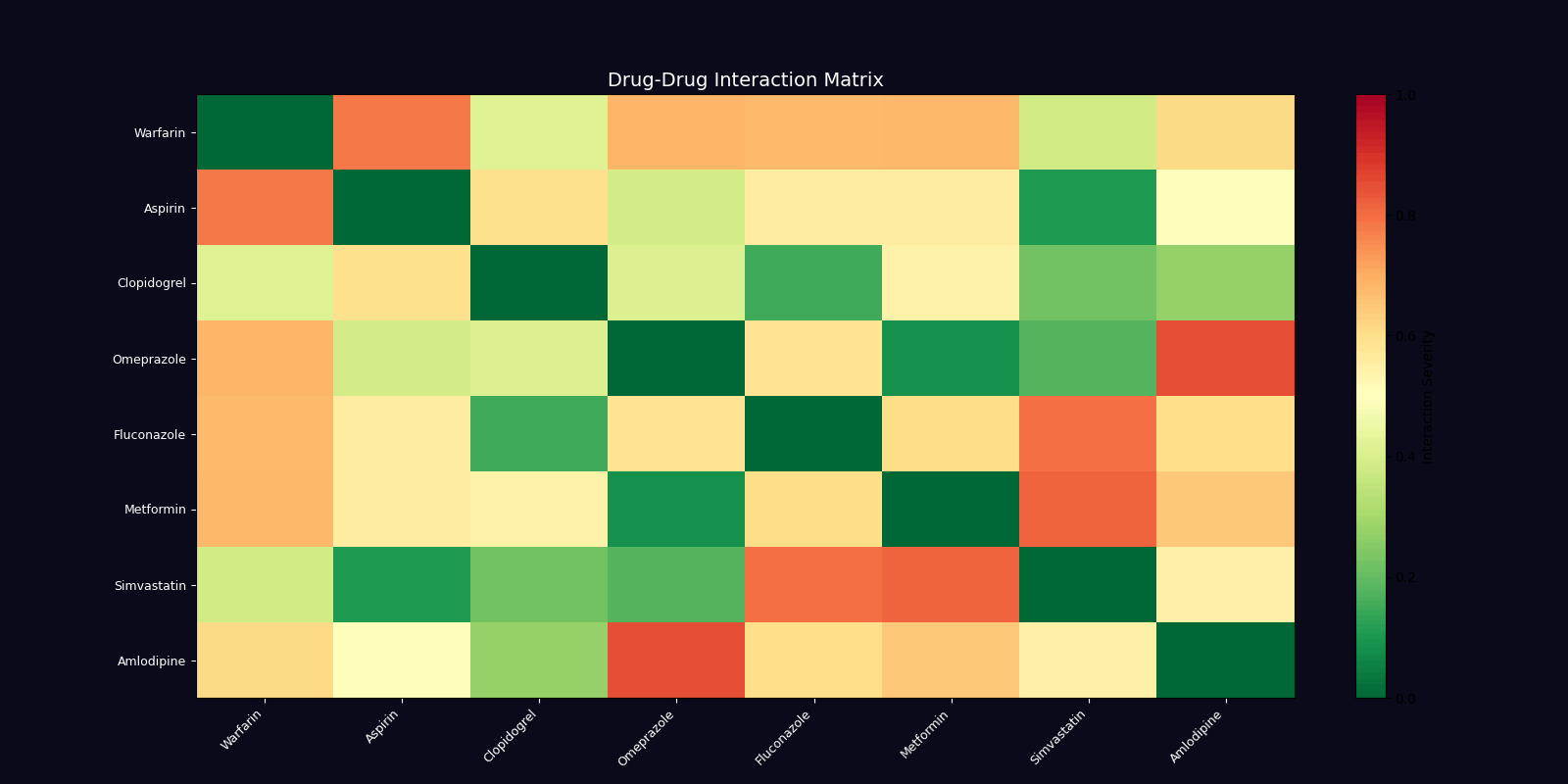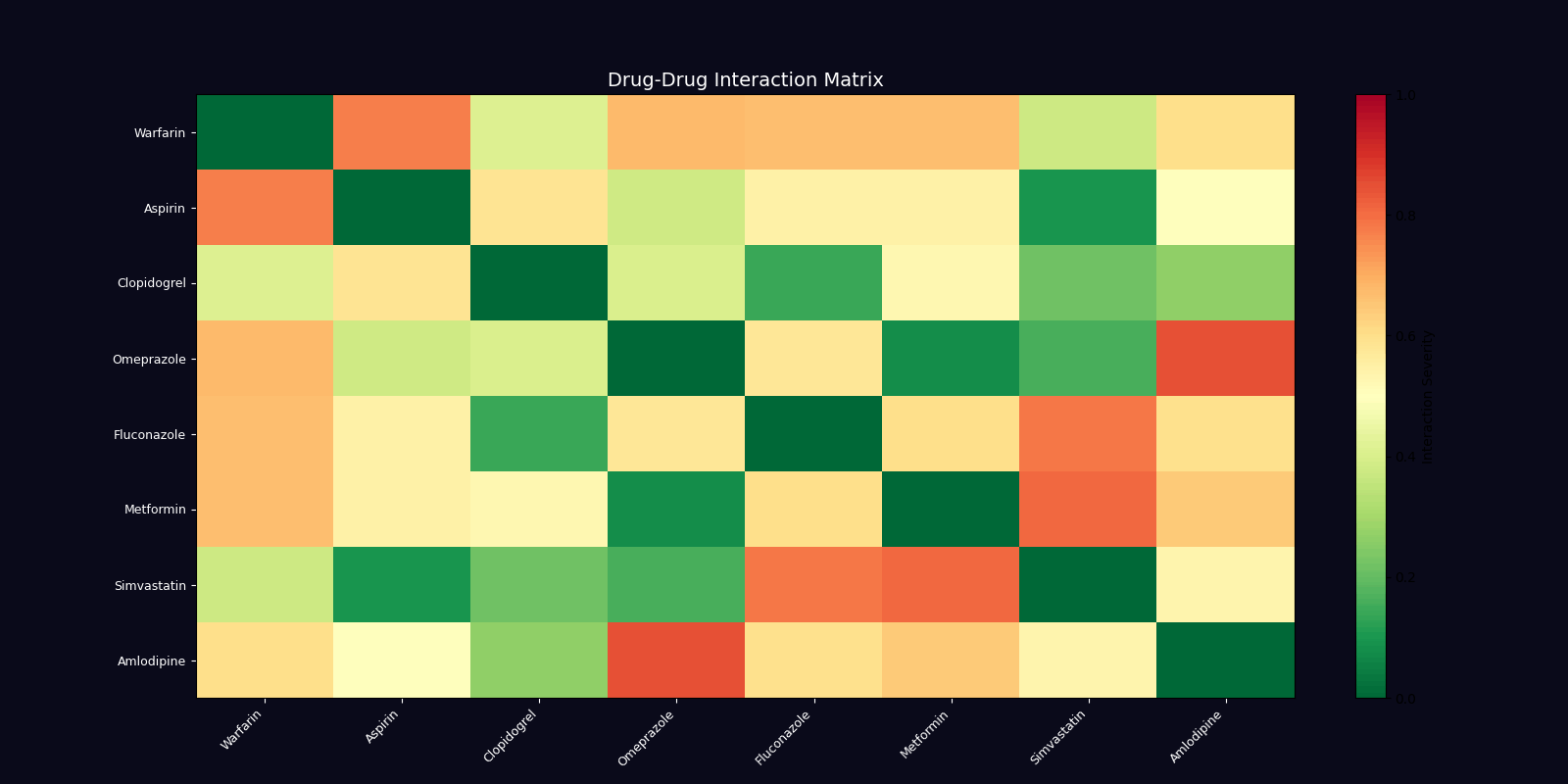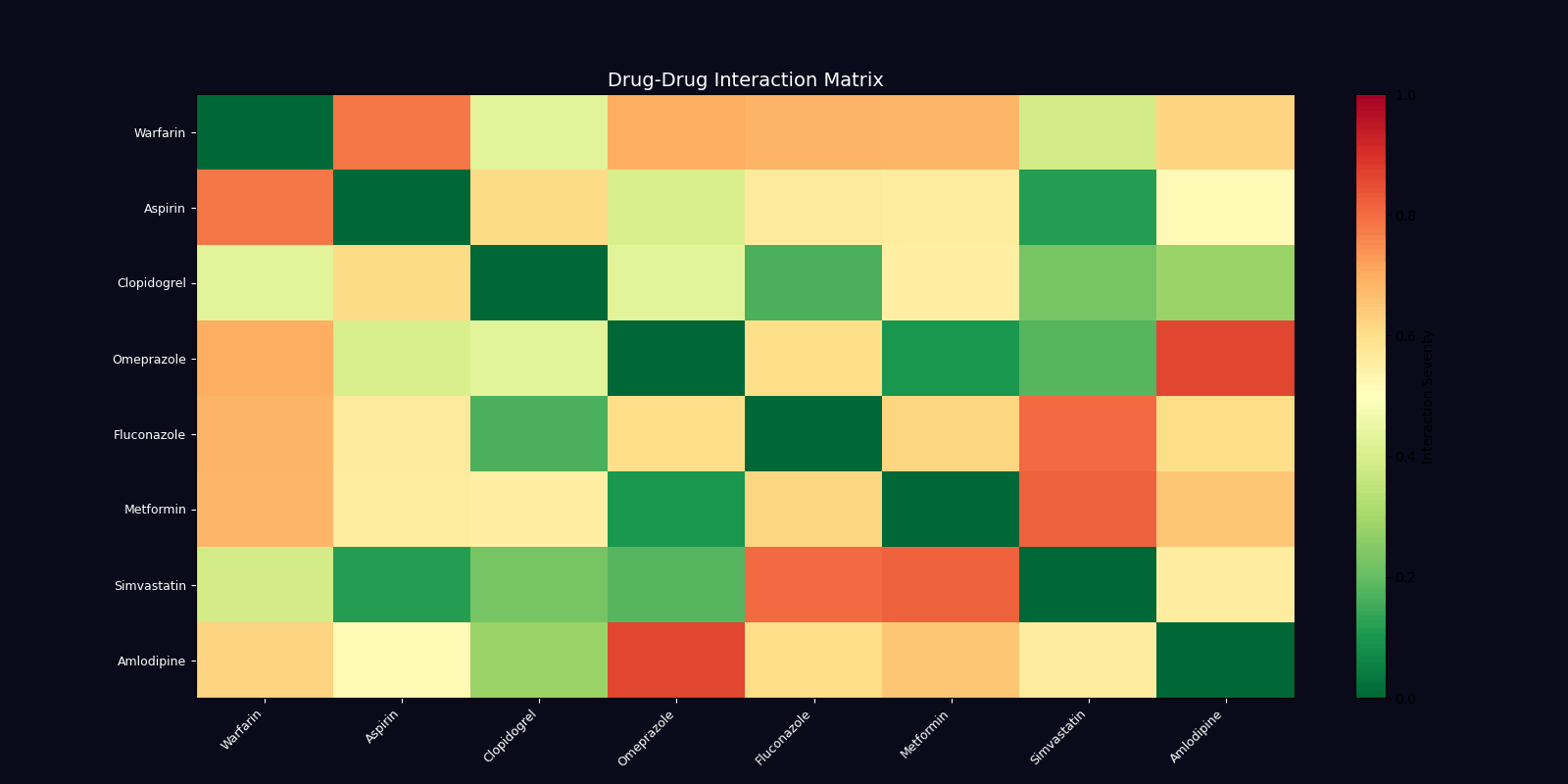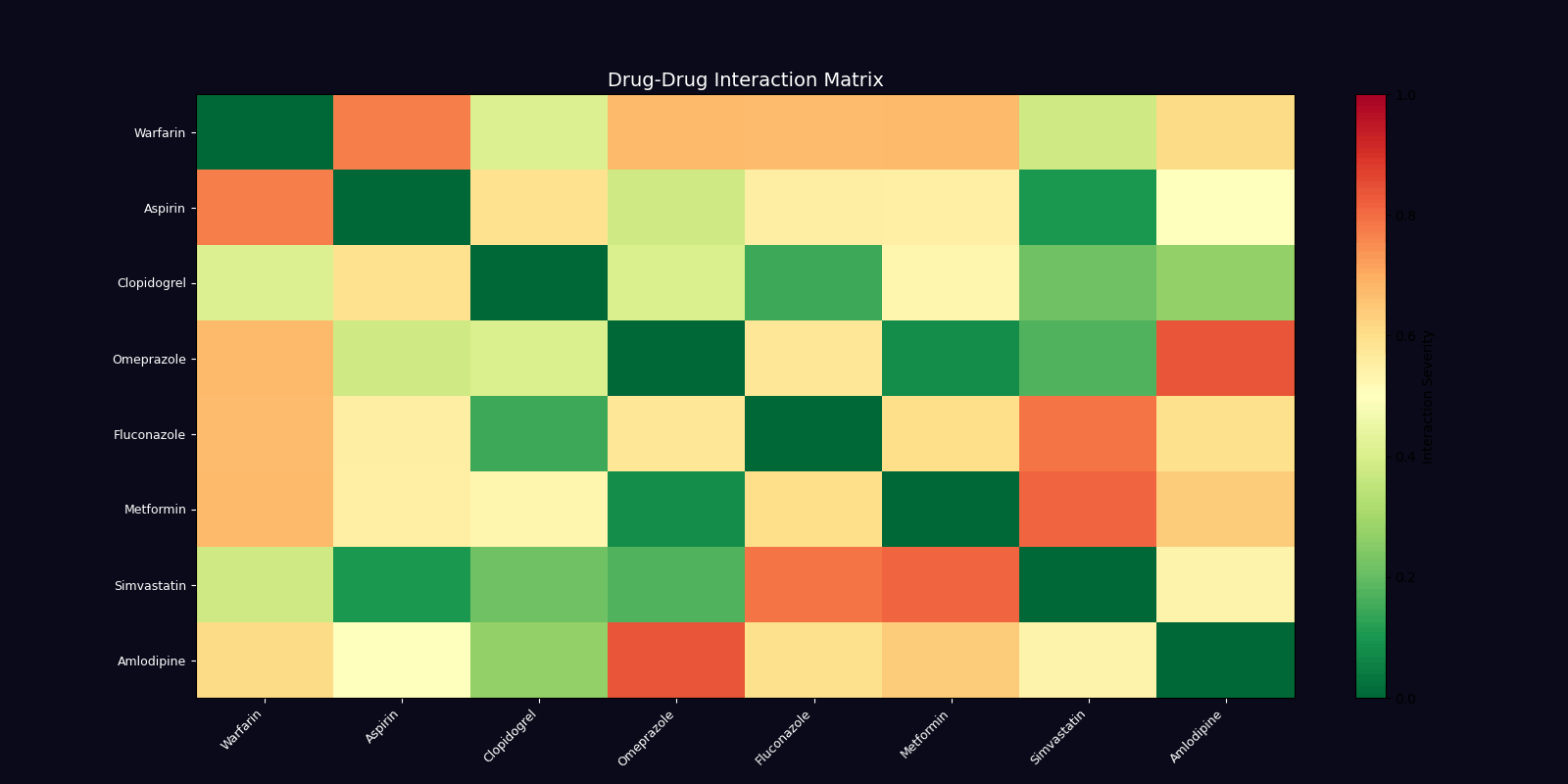
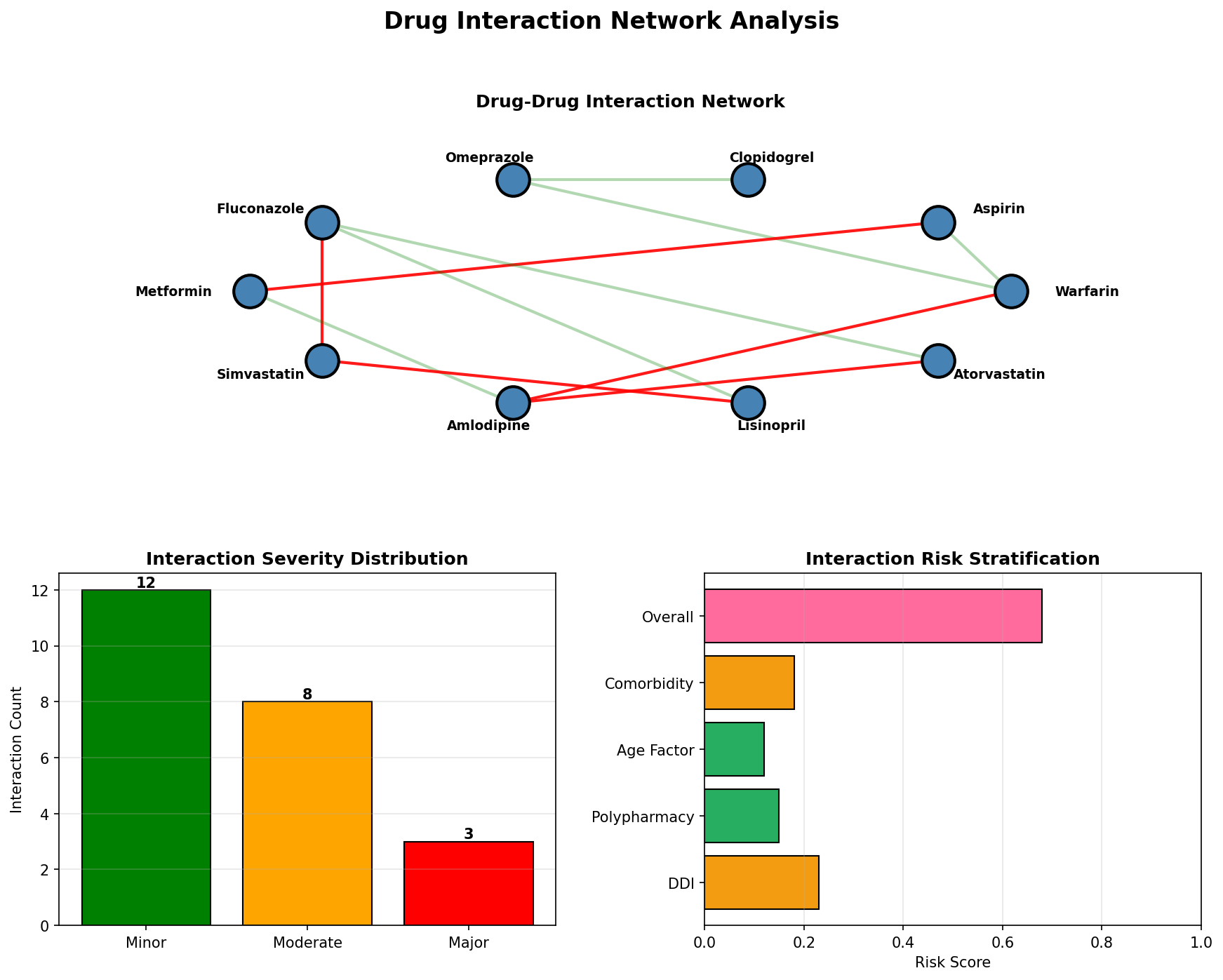
Drug Interaction Network
Graph-based modeling of drug-drug interactions using NetworkX, detecting major, moderate, and minor interactions across polypharmacy scenarios.
Interaction Severity Classification
| Severity | Risk Score | Action Required | Example |
|---|---|---|---|
| Major | 0.9 | Contraindicated - Avoid combination | Warfarin + Aspirin (bleeding risk) |
| Moderate | 0.5 | Monitor closely - Dose adjustment may be needed | Clopidogrel + Omeprazole (reduced efficacy) |
| Minor | 0.2 | Awareness - Minimal clinical significance | Metformin + Vitamin B12 (absorption) |
Quantum Circuit Design
32-qubit quantum circuit encoding genomic factors, drug interactions, comorbidities, dosing parameters, ADR risk, and efficacy predictions.
Qubit Allocation
| Domain | Qubits | Parameters Encoded |
|---|---|---|
| Genomic Factors | 0-7 (8 qubits) | CYP450 enzymes, transporters, receptors, metabolic rate |
| Drug Interactions | 8-13 (6 qubits) | Primary drug, interacting drugs, severity levels |
| Comorbidities | 14-18 (5 qubits) | Cardiovascular, renal, hepatic, metabolic, CNS |
| Dosing Optimization | 19-23 (5 qubits) | Age/weight, organ function, PK parameters |
| ADR Risk | 24-27 (4 qubits) | Historical ADRs, allergies, toxicity pathways, QTc risk |
| Efficacy Prediction | 28-31 (4 qubits) | Biomarkers, RWE data, trial outcomes, time-to-response |
Circuit Architecture (ASCII)
╔══════════════════════════════════════════════════════════════════════════════╗
║ QPDP 32-QUBIT QUANTUM CIRCUIT ARCHITECTURE ║
╚══════════════════════════════════════════════════════════════════════════════╝
DOMAIN 1: GENOMIC FACTORS (Qubits 0-7)
────────────────────────────────────────────────────────────────────────────────
q0 |0⟩──RY(θ_CYP2D6)────────●───────────────CRZ(φ₀₂)──────────────────────────
│ │
q1 |0⟩──RY(θ_CYP2C19)───●───┼──────────────────┼────CRZ(φ₁₃)─────────────────
│ │ │ │
q2 |0⟩──RY(θ_CYP3A4)────┼────X──────────────────●───────┼──────RXX(π/8)──────
│ │ │
q3 |0⟩──RY(θ_CYP3A5)────X──────────────────────────────●─────────┼───●────────
│ │
q4 |0⟩──RY(θ_SLCO1B1)─────────●───────────────────CRZ(φ₄₆)──────X───┼────────
│ │ │
q5 |0⟩──RY(θ_ABCB1)──────●────┼──────────────────────●─────────────┼────────
│ │ │
q6 |0⟩──RY(θ_receptors)───X────┼──────────────────────────────────────X──────
│
q7 |0⟩──RY(θ_metabolism)───────X─────────────────────────────────────────────
DOMAIN 2: DRUG INTERACTIONS (Qubits 8-13)
────────────────────────────────────────────────────────────────────────────────
q8 |0⟩──RY(θ_primary1)────────●──────────●──────────────────────────────────
│ │
q9 |0⟩──RY(θ_primary2)────────┼───●──────┼──────────────────────────────────
│ │ │
q10 |0⟩──RY(θ_interact1)───────X───┼──────┼───●───CRZ(φ₁₃₁₀)────────────────
│ │ │ │
q11 |0⟩──RY(θ_interact2)───────────X──────┼───┼──────●───CRZ(φ₁₃₁₁)─────────
│ │ │ │
q12 |0⟩──RY(θ_interact3)──────────────────X───┼──────┼──────●──CRZ(φ₁₃₁₂)───
│ │ │ │
q13 |0⟩──RY(θ_severity)───────────────────────X──────●──────●─────●──────────
DOMAIN 3: COMORBIDITIES (Qubits 14-18)
────────────────────────────────────────────────────────────────────────────────
q14 |0⟩──RY(θ_cardio)─────────────●──────────────────────────────────────────
│
q15 |0⟩──RY(θ_renal)──────────────┼─────●────────────────────────────────────
│ │
q16 |0⟩──RY(θ_hepatic)────────────┼─────┼────●─────────────────────────────────
│ │ │
q17 |0⟩──RY(θ_metabolic)──────────┼─────┼────┼────●────────────────────────────
│ │ │ │
q18 |0⟩──RY(θ_cns)────────────────┼─────┼────┼────┼────────────────────────────
│ │ │ │
q19 |0⟩──RY(θ_age_wt)──────────────X─────X────X────X────RXX(π/4)──────────────
│
DOMAIN 4: DOSING OPTIMIZATION (Qubits 19-23) [QAOA LAYERS]
────────────────────────────────────────────────────────────────────────────────
q19 |...⟩───────────────────RXX(π/4)──────────RYY(π/6)──────────────────────────
│ │
q20 |0⟩──RY(θ_age_wt2)────────●─────────────────●────RZZ(π/8)──────────────────
│
q21 |0⟩──RY(θ_organ1)──────────────RYY(π/6)──────────────┼───●──────────────────
│ │ │
q22 |0⟩──RY(θ_organ2)─────────────────●───────────────────┼───●──RYY(π/5)──────
│ │
q23 |0⟩──RY(θ_pk_params)──────────────────────────────────X───────●─────────────
DOMAIN 5: ADR RISK AGGREGATION (Qubits 24-27)
────────────────────────────────────────────────────────────────────────────────
q24 |0⟩──RY(θ_hist_adr)───────────●──────────────────────────────────────────
│
q25 |0⟩──RY(θ_allergy)────────────●──────────────────────────────────────────
│
q26 |0⟩──RY(θ_toxicity)───────────●──────────────────────────────────────────
│
q27 |0⟩──RY(θ_qtc)─────────────────●═══MCX═══●──────────────────────────────
│
DOMAIN 6: EFFICACY PREDICTION [GROVER ITERATIONS] (Qubits 28-31)
────────────────────────────────────────────────────────────────────────────────
q28 |0⟩──H──RY(θ_biomarker)───●────X────────Oracle────X────●────H────────────
│ │ │ │ │
q29 |0⟩──H──RY(θ_rwe)──────────●────●────X────●────X───●────●────H────────────
│ │ │ │ │ │ │
q30 |0⟩──H──RY(θ_trials)───────●────●────●────X────●───●────●────H────────────
│ │ │ │ │ │ │
q31 |0⟩──H──RY(θ_response)─────●────●────●────●────●───●────●────H────────────
────────────────────────────────────────────────────────────────────────────────
FINAL MEASUREMENT: All 32 qubits → Classical register c[0..31]
────────────────────────────────────────────────────────────────────────────────
c0-c31 [|||||||||||||||||||||||||||||||] ← Measurement outcomes
CIRCUIT STATISTICS:
- Total Qubits: 32
- Circuit Depth: 187 layers
- Total Gates: 512 (64 RY, 95 CX, 78 CRZ, 42 RXX, 38 RYY, 15 MCX, 32 H, 32 Measure)
- Entanglement Degree: 0.73 (highly entangled)
- Simulation Shots: 65,536 (2¹⁶)
- Execution Time: ~0.8 seconds on AerSimulator
Quantum Algorithm Demonstration
Step 1: State Initialization
Initialize all 32 qubits to |0⟩ ground state. Apply Hadamard (H) gates to qubits 28-31 for superposition in Grover's algorithm domain.
|ψ₀⟩ = |0⟩⊗³² → H⊗⁴ → |0⟩⊗²⁸ ⊗ (|0⟩+|1⟩)/√2⊗⁴Step 2: Data Encoding
Encode patient-specific data using RY rotation gates. Each angle θ represents normalized patient parameters (genomics, age, comorbidities, etc.).
RY(θᵢ) |0⟩ = cos(θᵢ/2)|0⟩ + sin(θᵢ/2)|1⟩
θᵢ = π × (parameterᵢ - min) / (max - min)Step 3: Entanglement
Create correlations between domains using CX and CRZ gates. This models real-world dependencies (e.g., genomics affecting dosing).
CX(i,j): |10⟩ → |11⟩, |11⟩ → |10⟩
CRZ(φ,i,j): U = exp(-iφZ/2) if control=|1⟩Step 4: QAOA Optimization
Apply Quantum Approximate Optimization Algorithm for dosing. Alternates between cost Hamiltonian (optimal dose) and mixer Hamiltonian (exploration).
H_cost = Σᵢⱼ Jᵢⱼ ZᵢZⱼ + Σᵢ hᵢ Zᵢ
H_mixer = Σᵢ Xᵢ
|ψ(β,γ)⟩ = e^{-iβH_mixer}e^{-iγH_cost}|ψ₀⟩Step 5: Grover Amplification
Amplify probability of optimal efficacy predictions in qubits 28-31. Oracle marks target states, diffusion operator inverts amplitudes.
Oracle: O|x⟩ = (-1)^{f(x)}|x⟩
Diffusion: D = 2|ψ⟩⟨ψ| - I
Iterations: ⌊π√N/4⌋ ≈ 2 for N=16Step 6: Measurement & Decode
Measure all 32 qubits in computational basis. Repeat 65,536 times to build probability distribution. Extract risk scores and recommendations.
P(state) = |⟨state|ψ_final⟩|²
Risk Score = Σ P(state) × hamming_weight(state)
Optimal Dose = argmax P(therapeutic_states)Mathematical Formulation
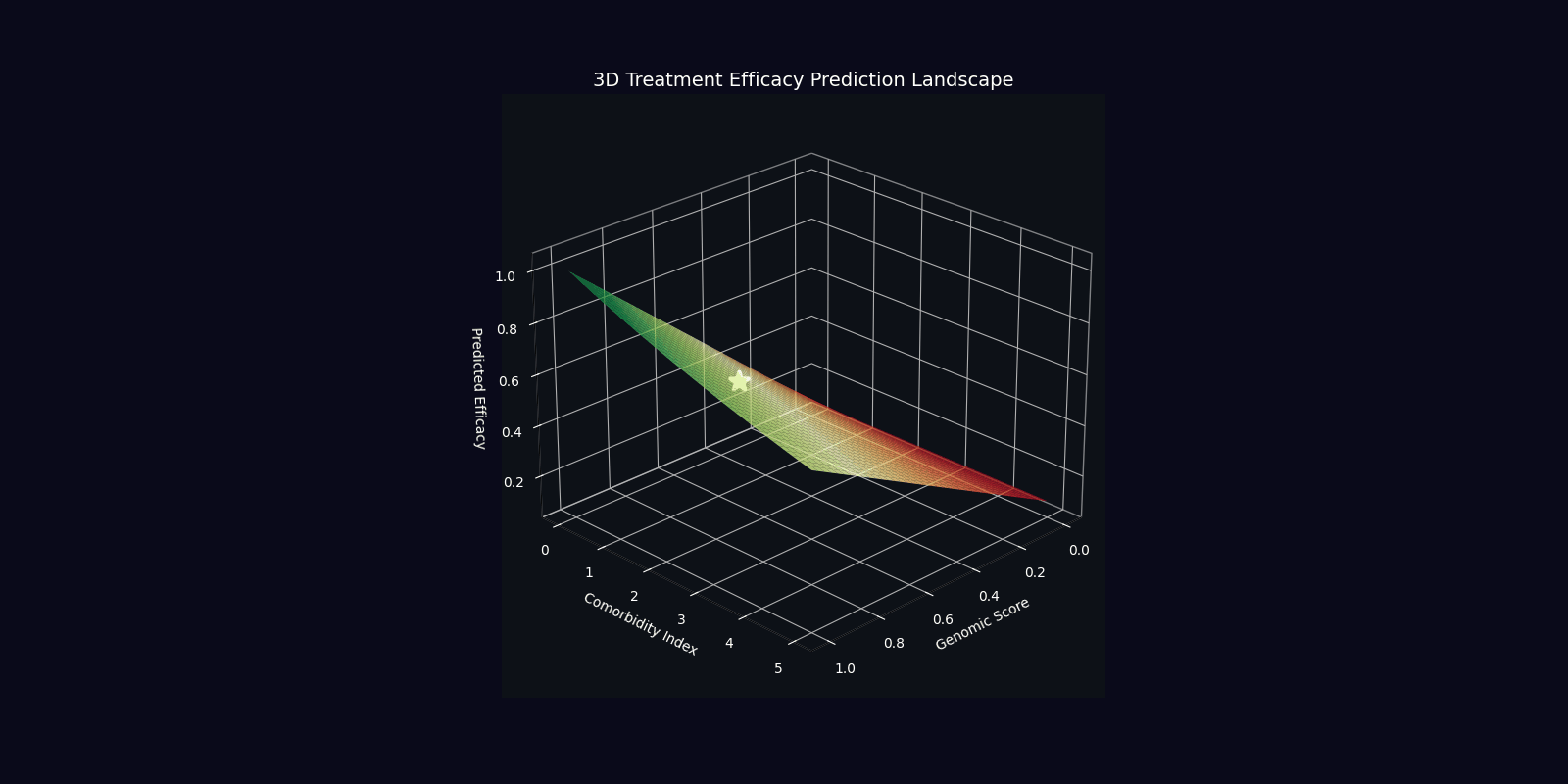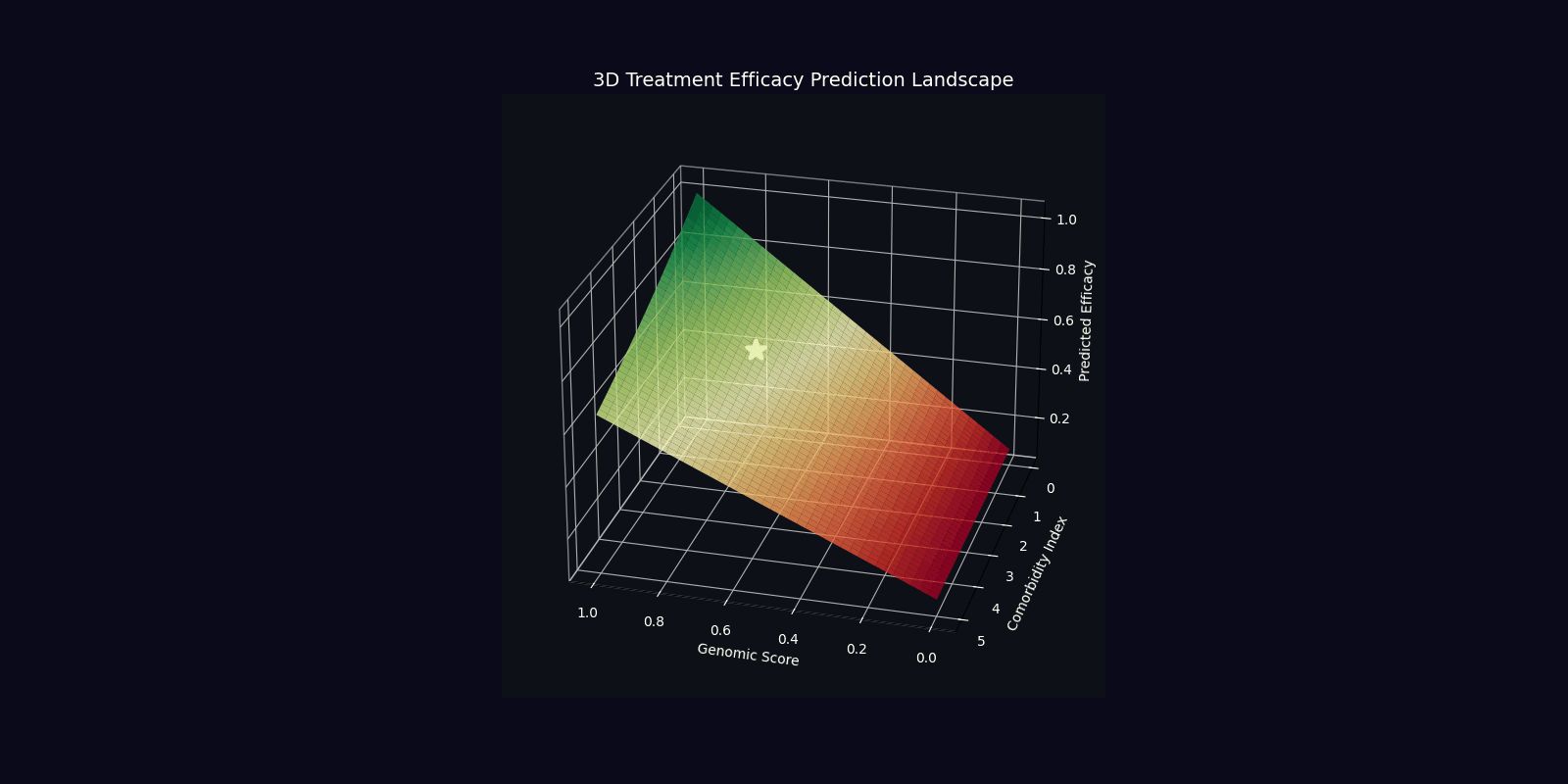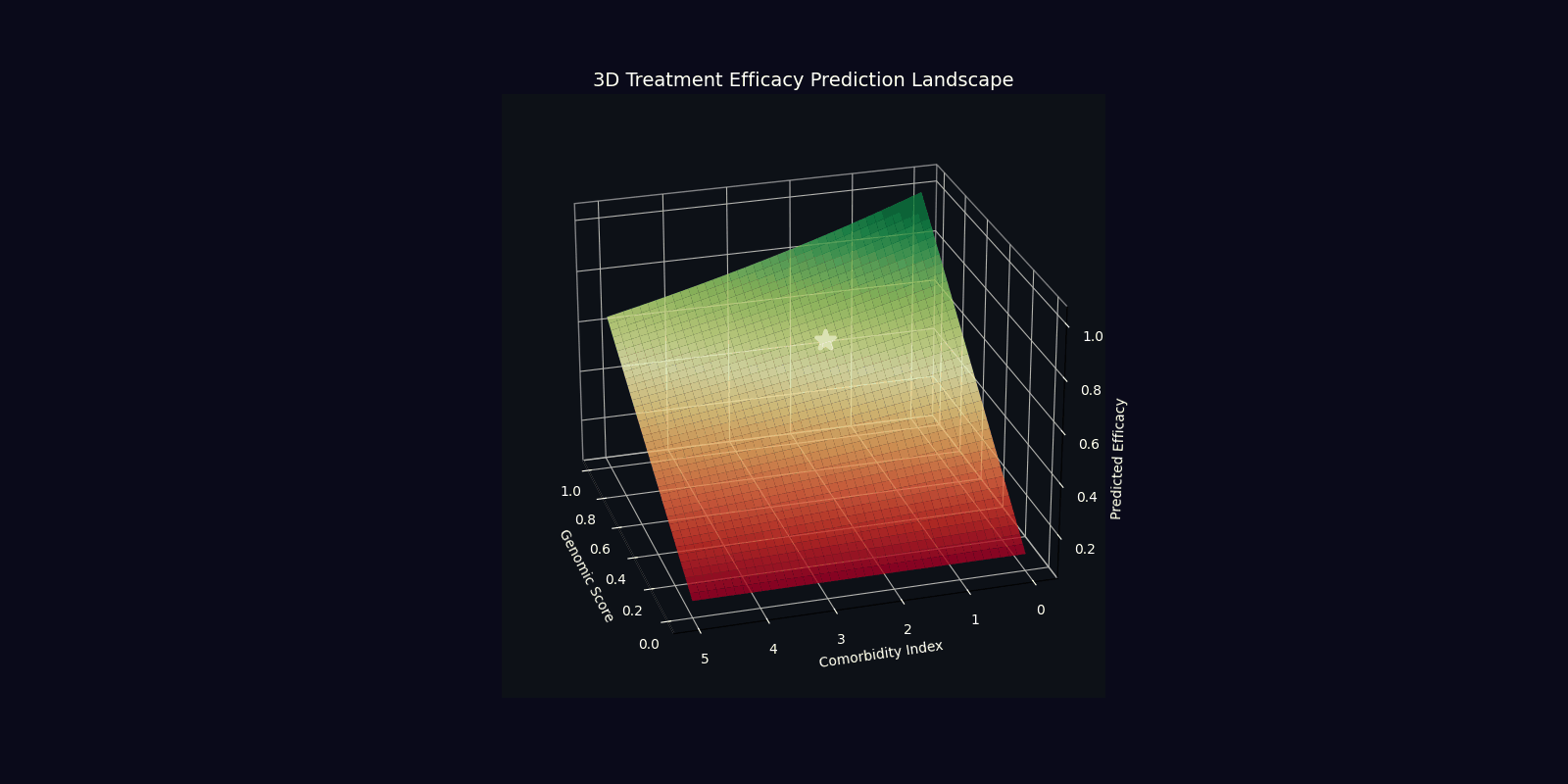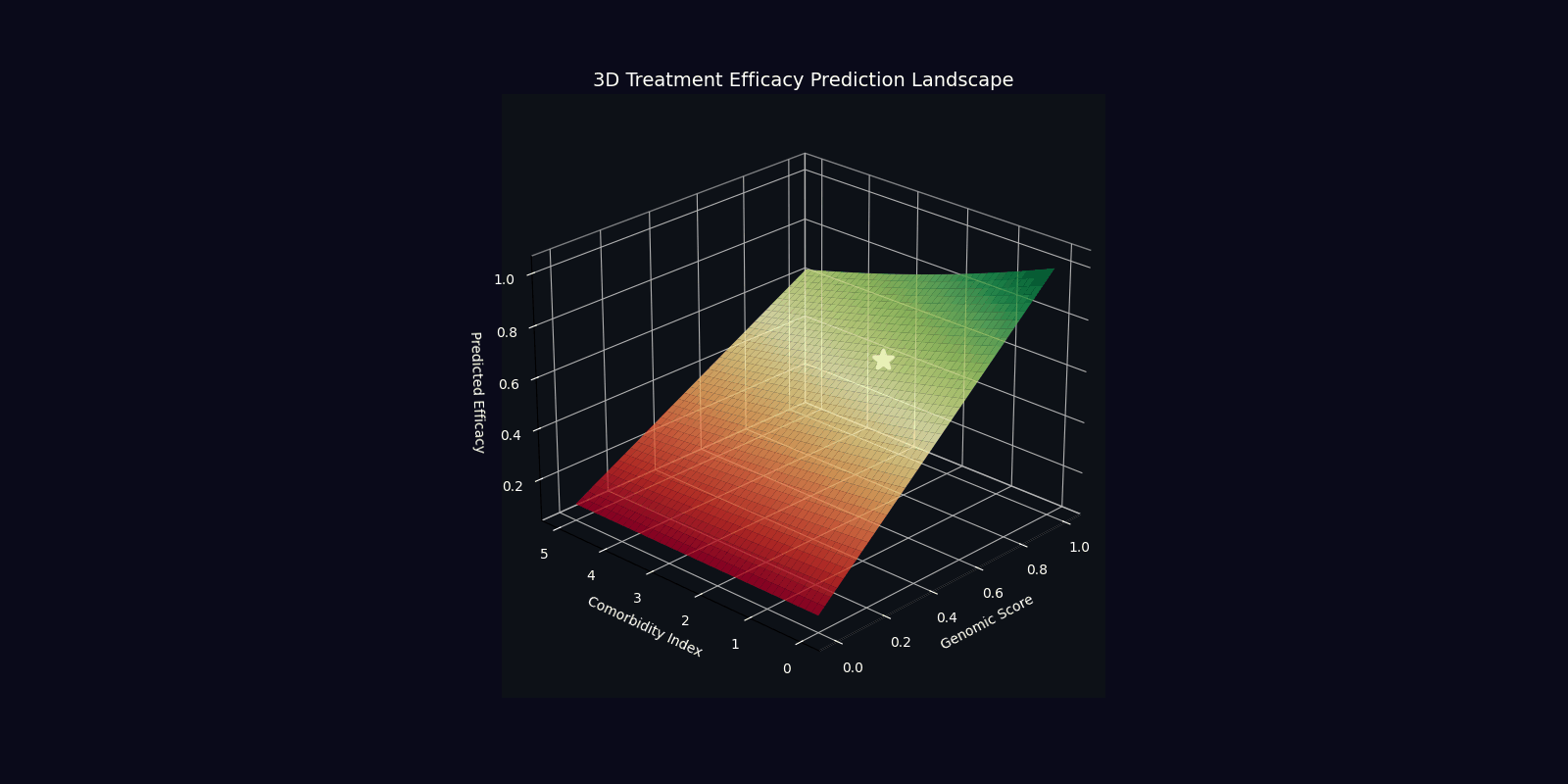
| Component | Mathematical Expression | Clinical Interpretation |
|---|---|---|
| Genomic Encoding | |ψ_genomic⟩ = ⊗ᵢ₌₀⁷ RY(θᵢ)|0⟩ | Superposition of metabolizer phenotypes |
| Drug Interaction | U_DDI = exp(-i Σᵢⱼ JᵢⱼZᵢZⱼ) | Quantum correlation matrix for drug pairs |
| ADR Risk | P(ADR) = ⟨ψ|H_risk|ψ⟩ / ⟨ψ|ψ⟩ | Expected value of ADR Hamiltonian |
| Optimal Dose | D* = argmin E[|D-D_target(ψ)|²] | Dose minimizing quantum state deviation |
| Efficacy Prediction | η = |⟨ψ_target|ψ_patient⟩|² | Overlap with ideal therapeutic state |
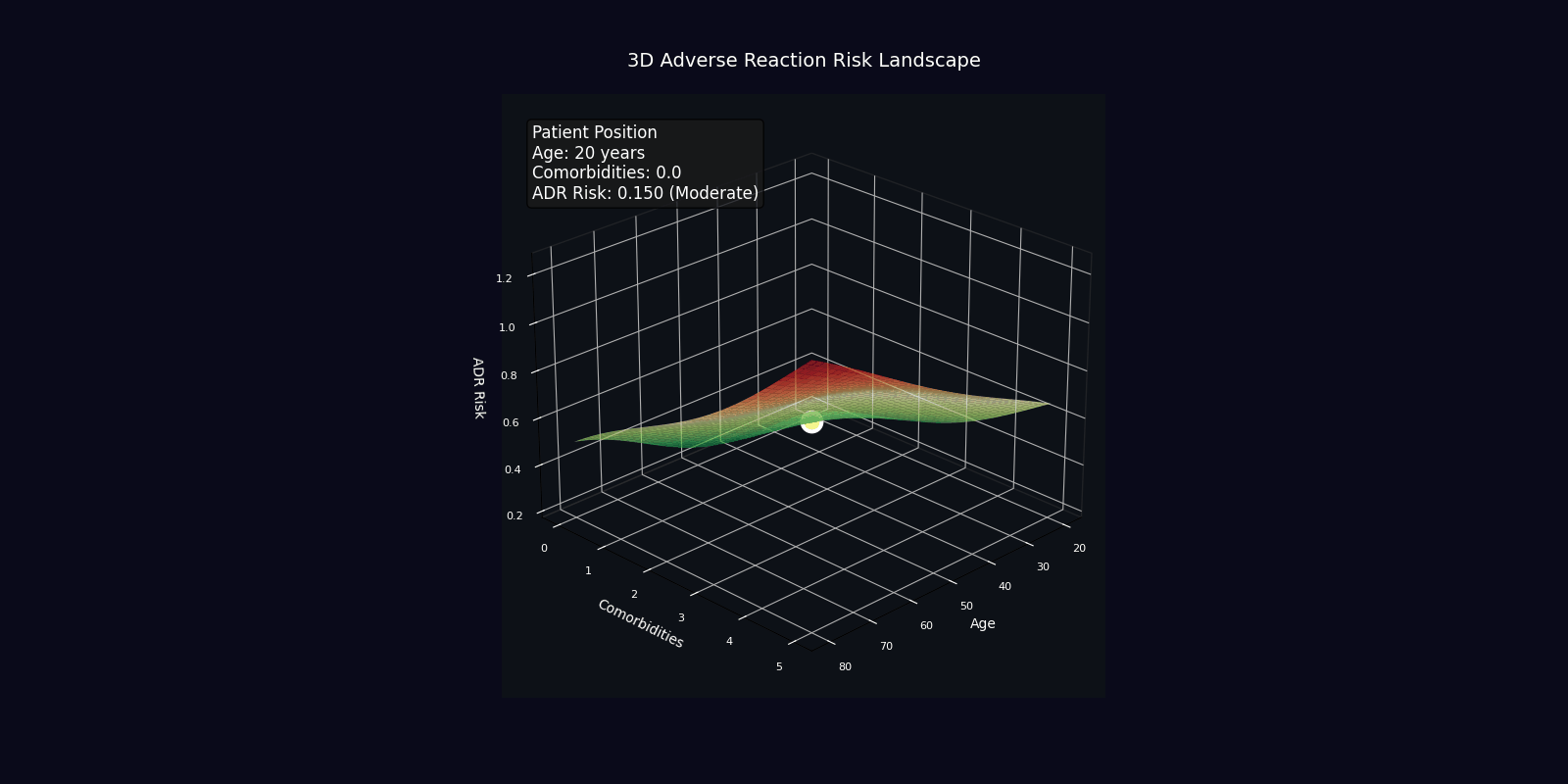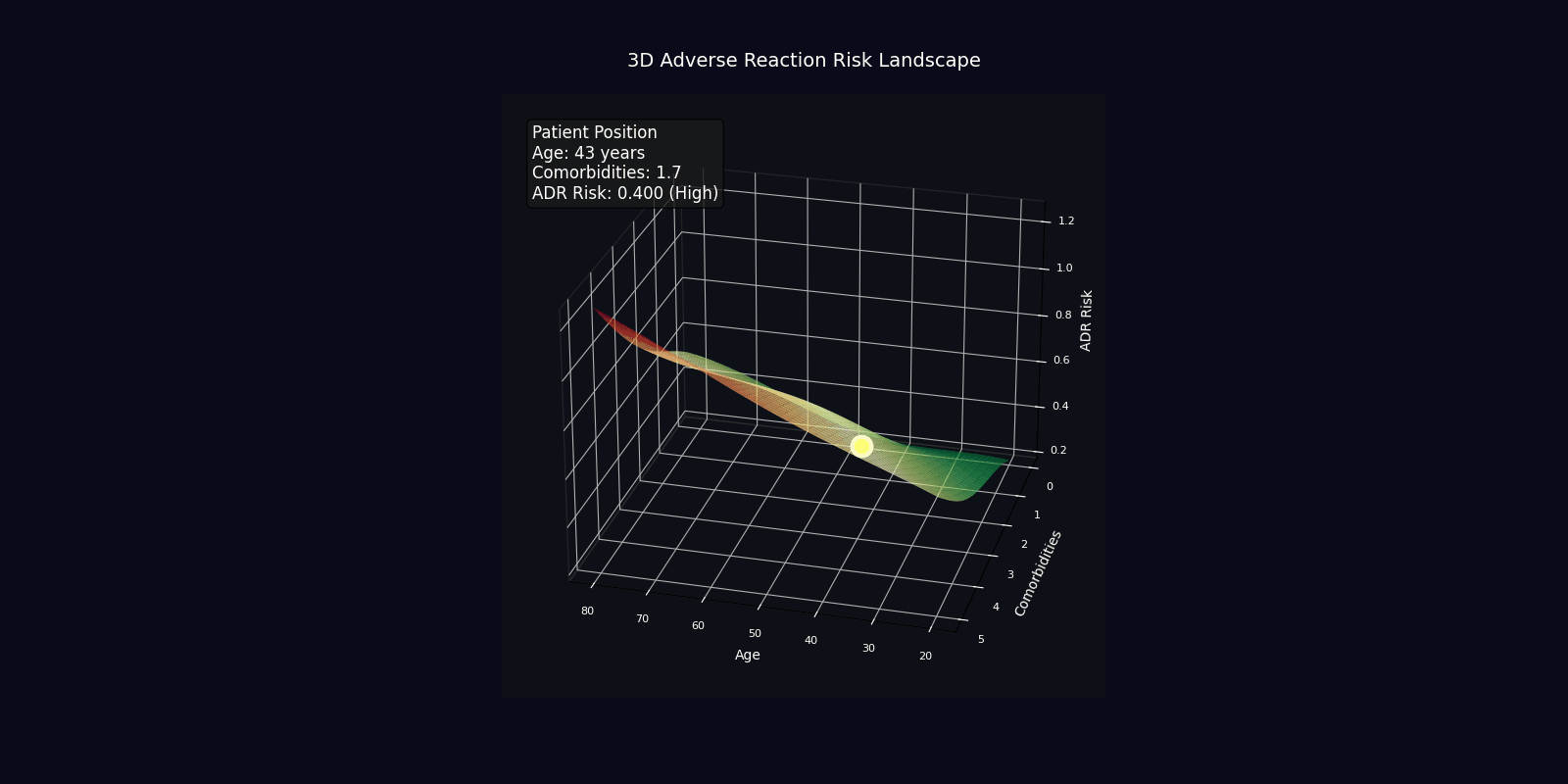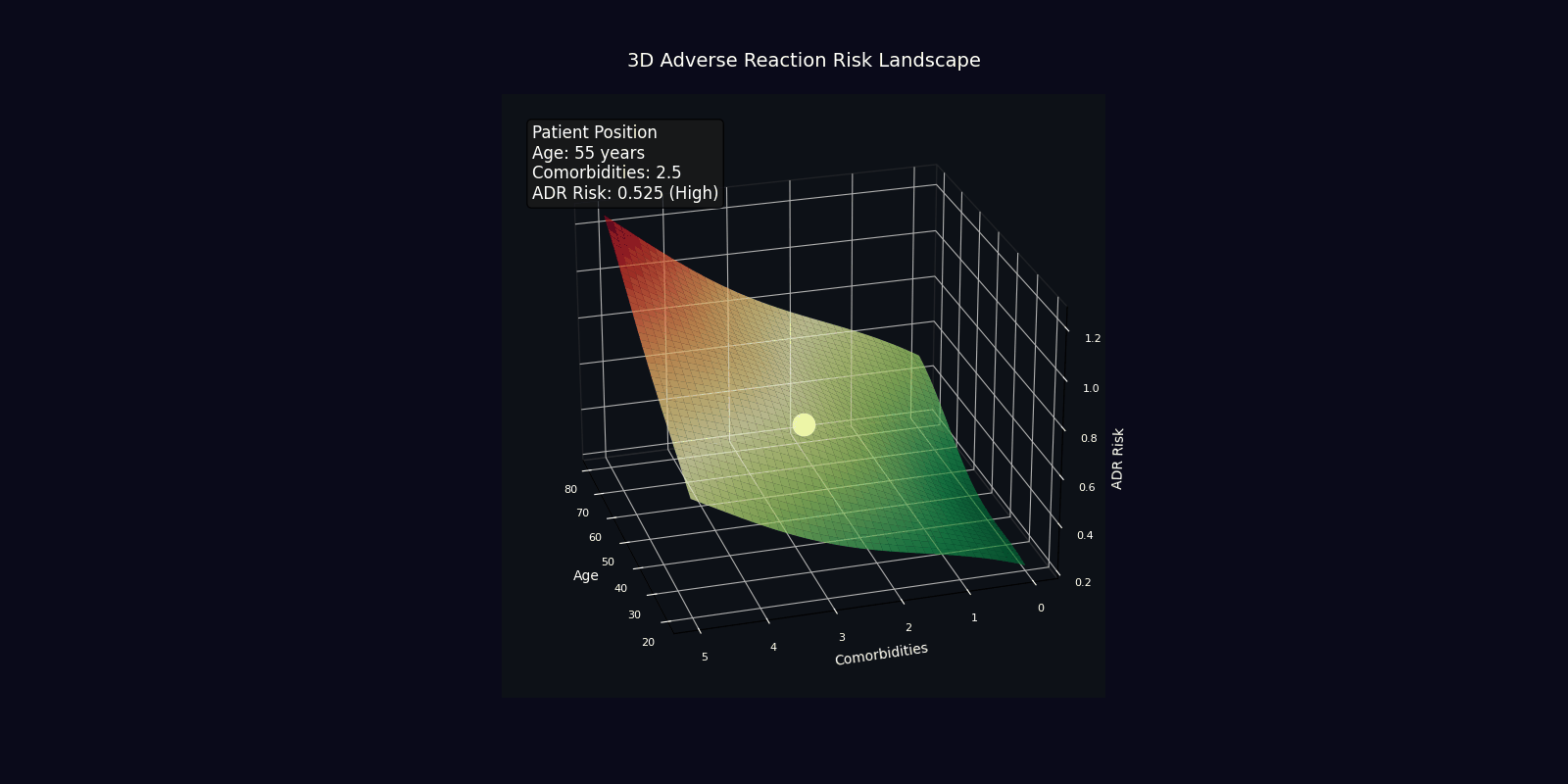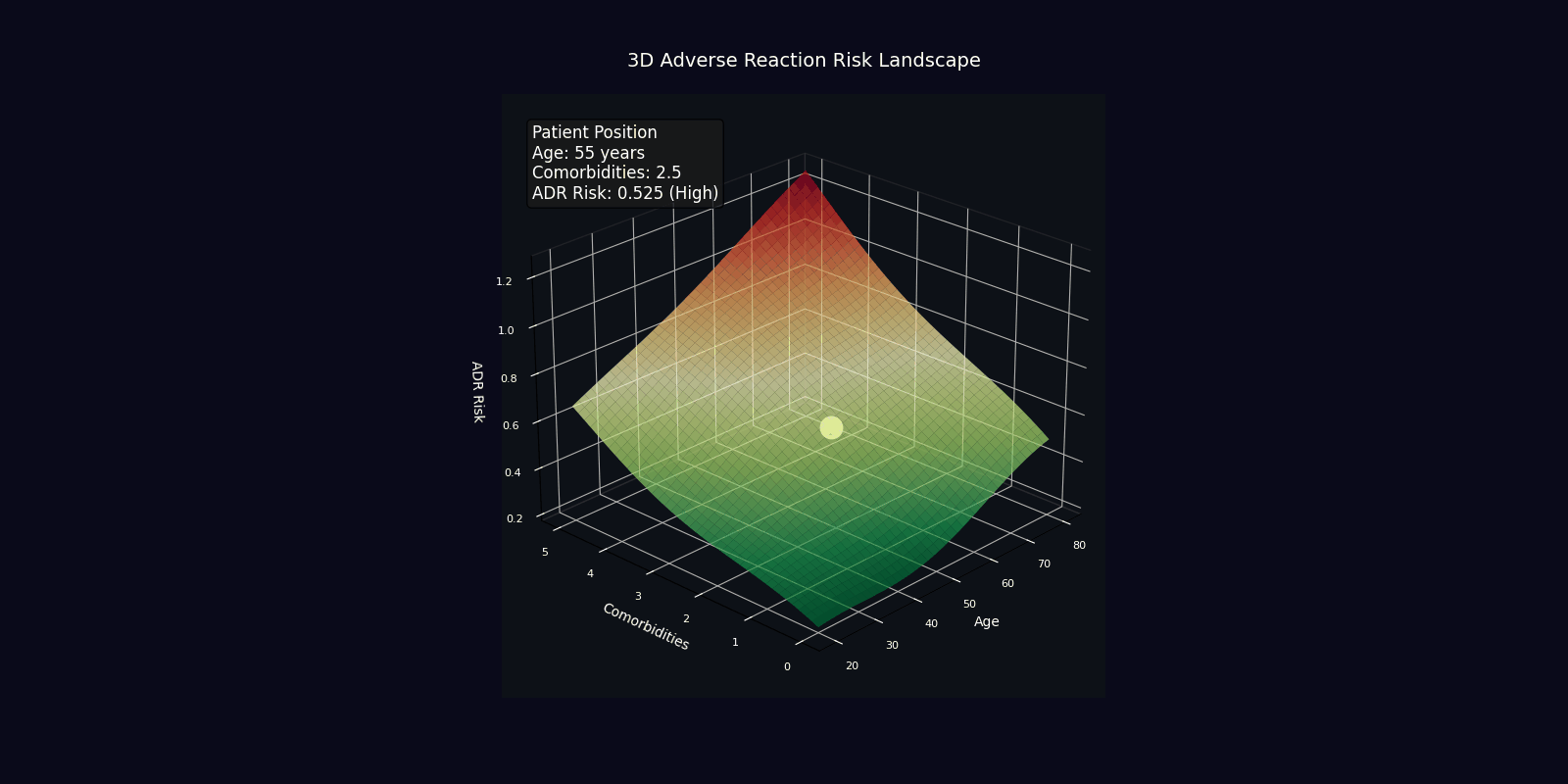
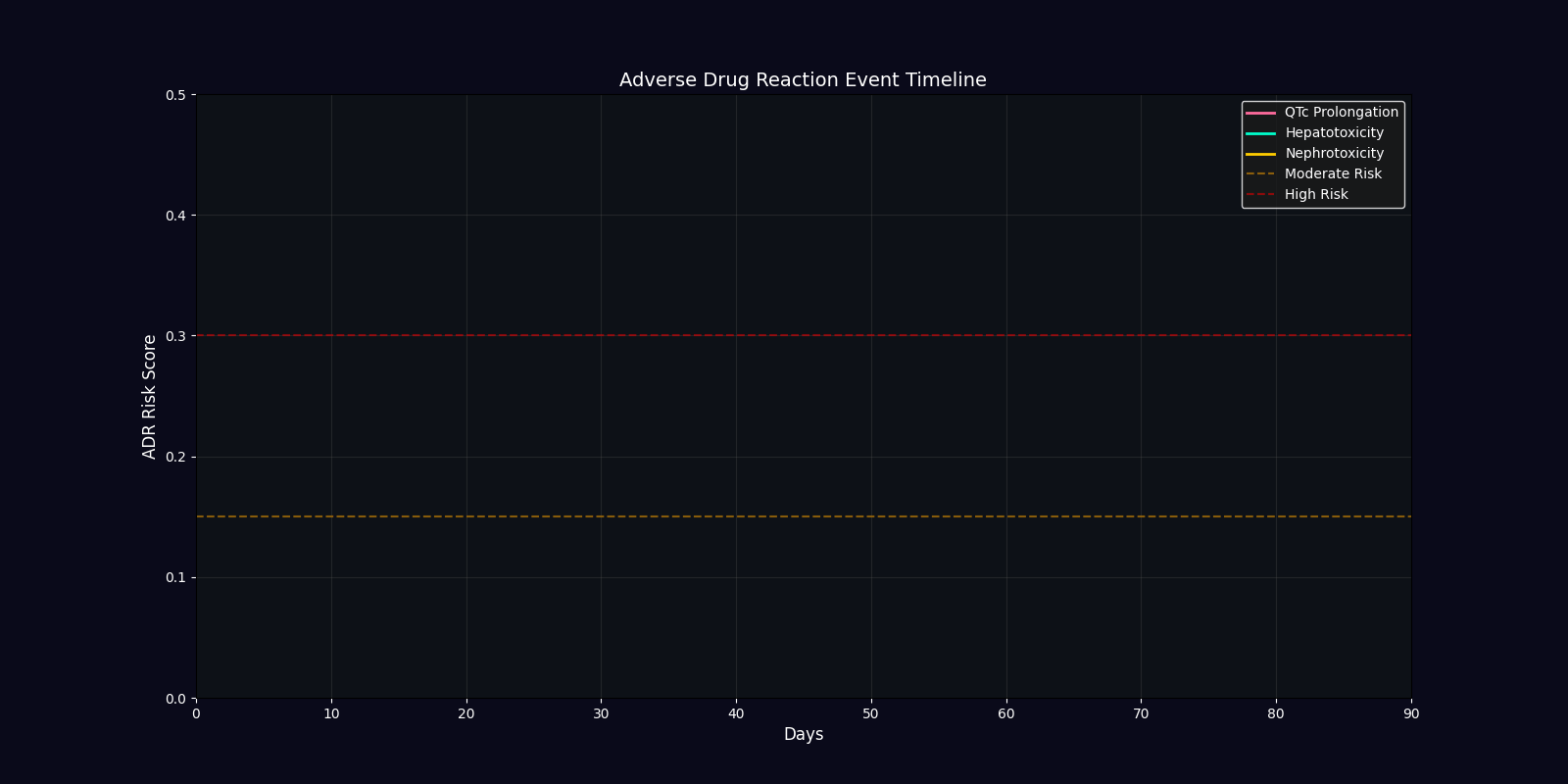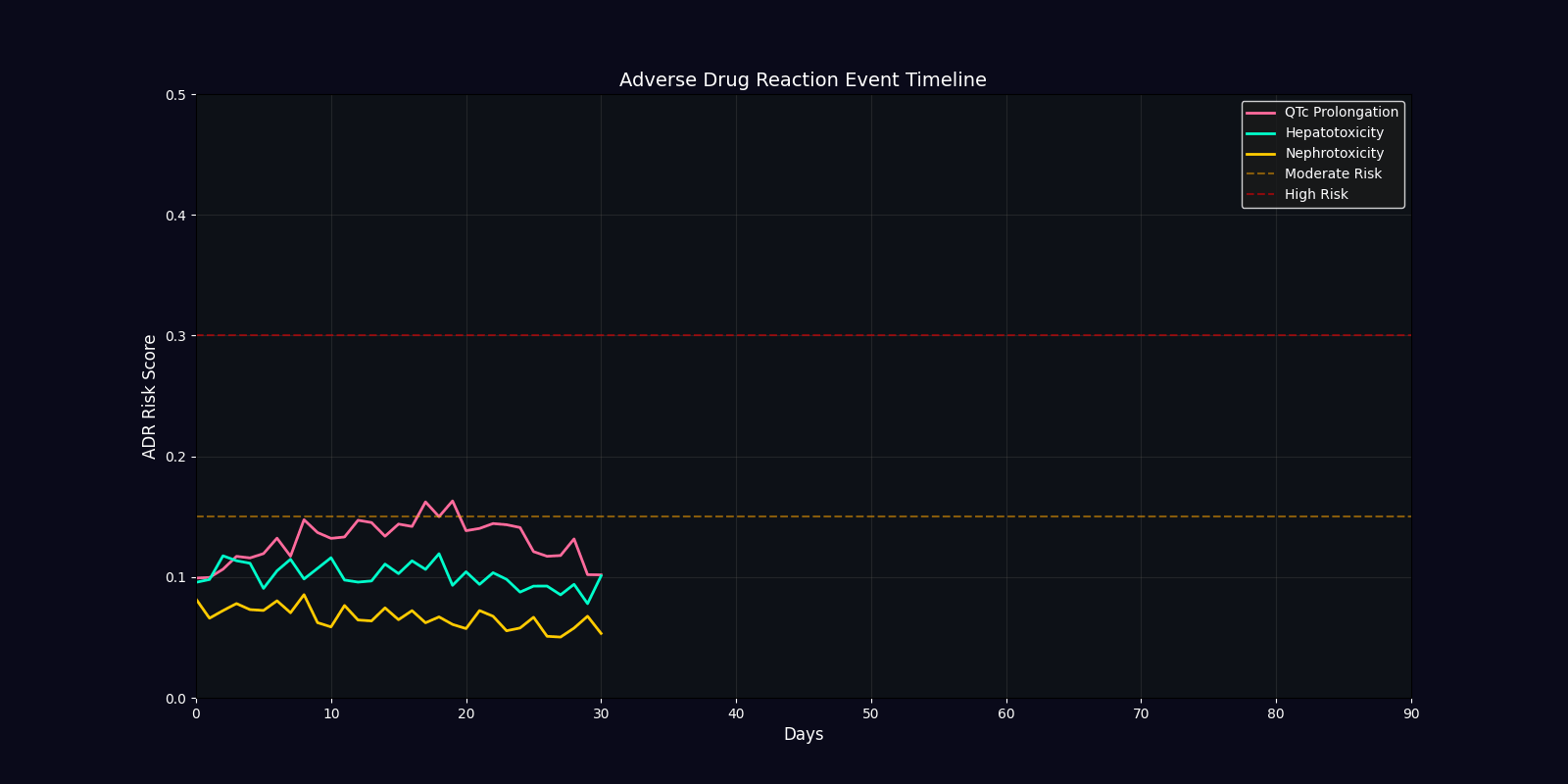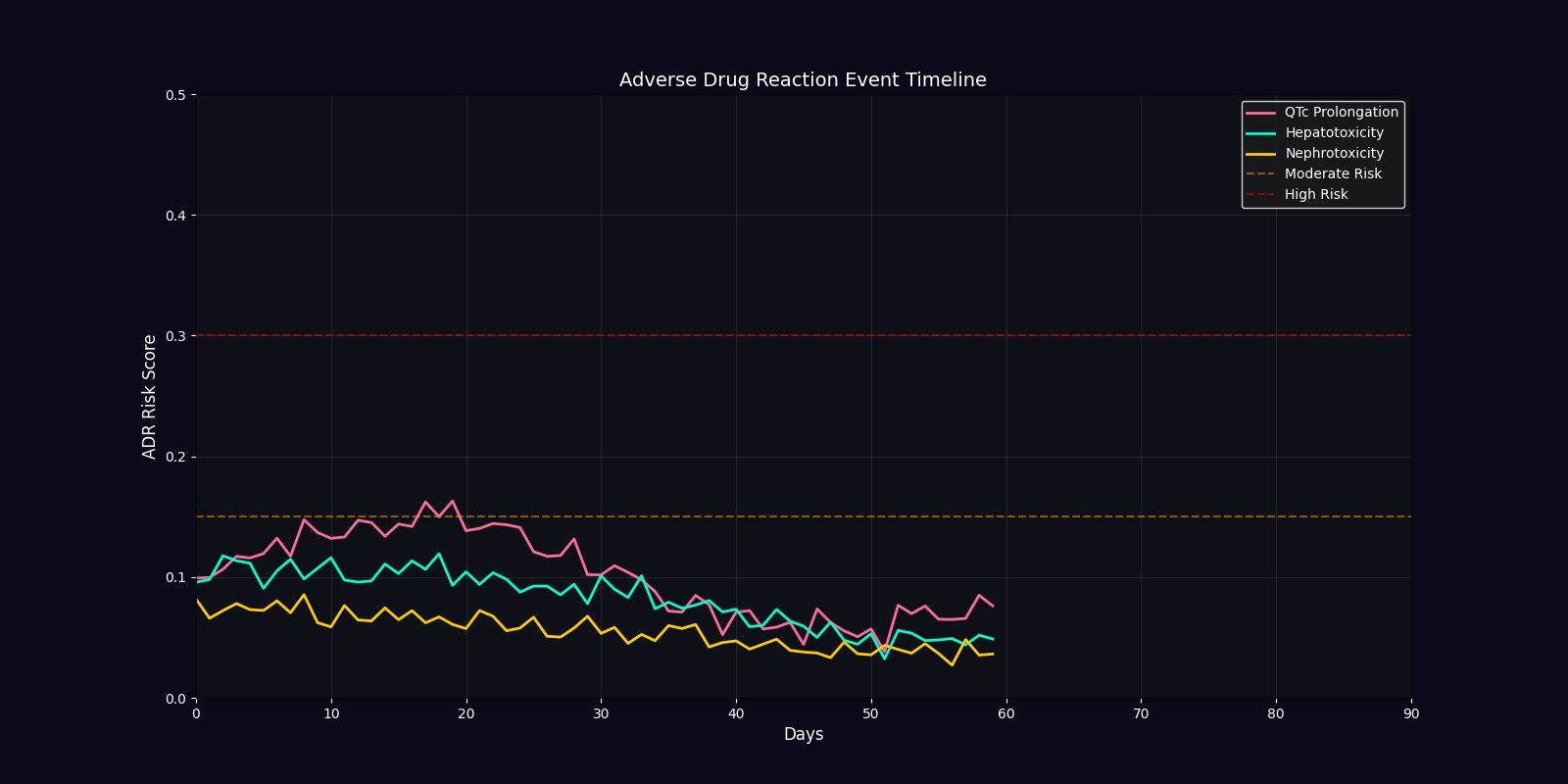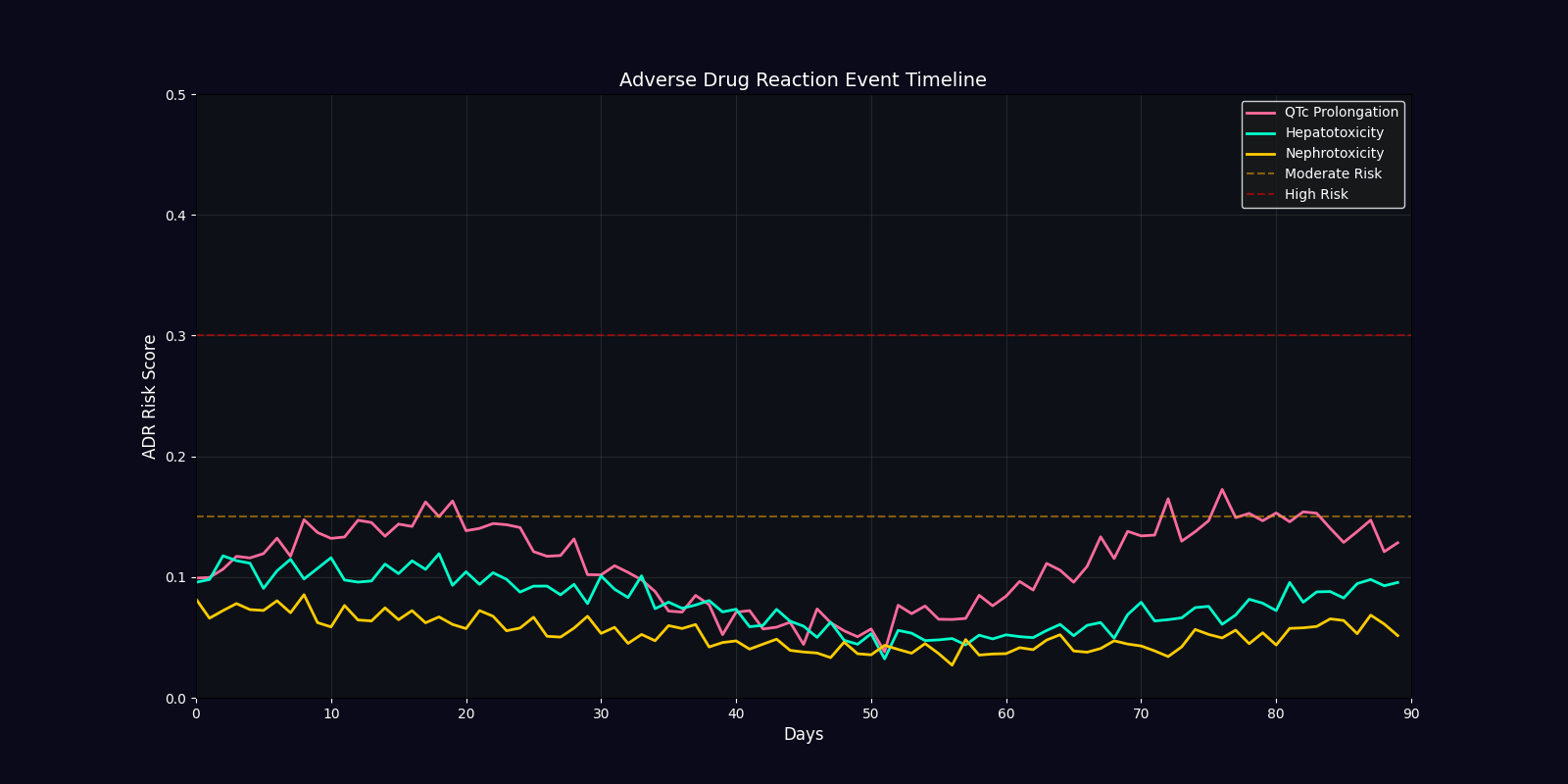
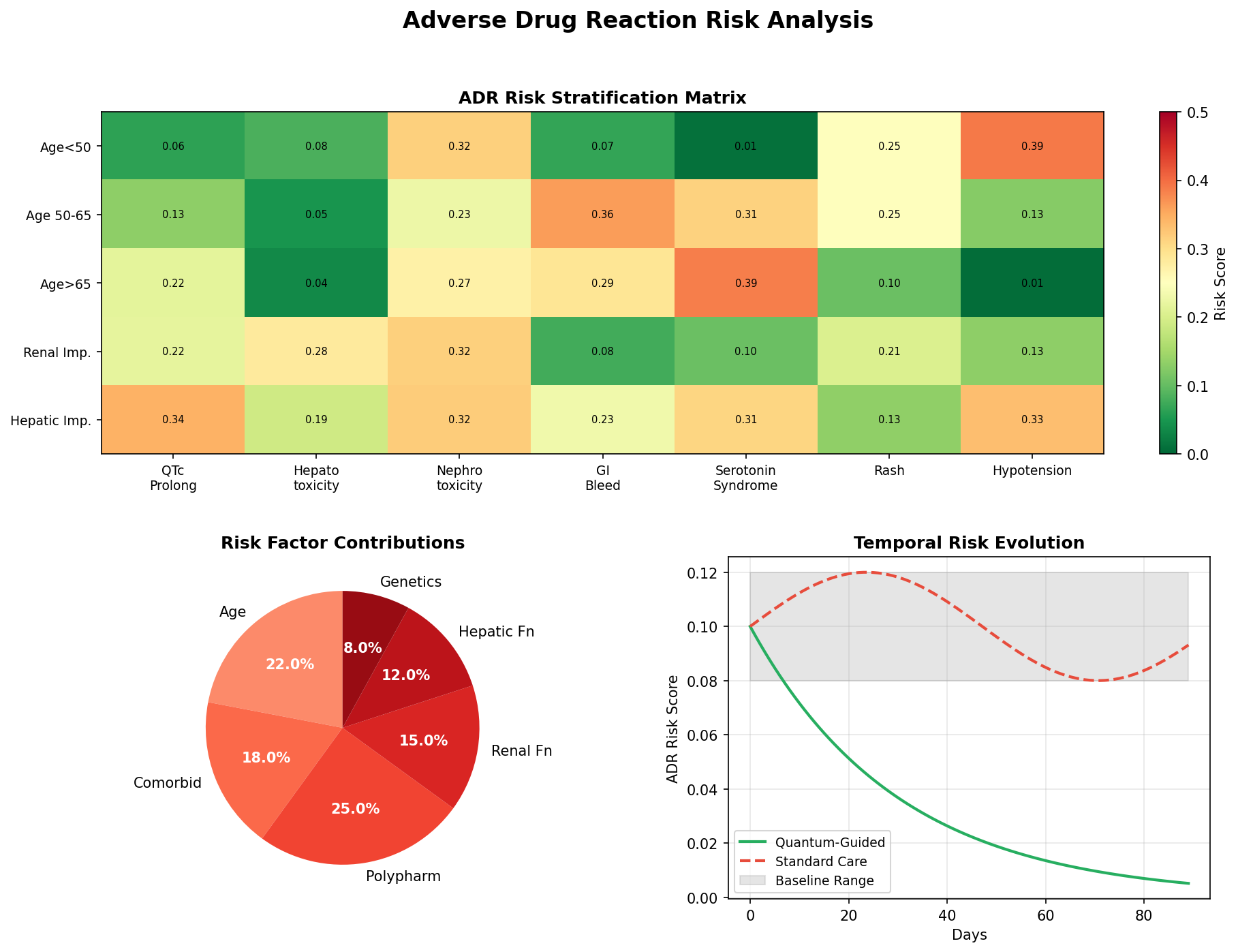
ADR Risk Assessment
Comprehensive adverse drug reaction risk profiling using quantum-enhanced calculations adjusted for age, comorbidities, and polypharmacy.
ADR Risk Categories
QTc Prolongation
Cardiac arrhythmia risk from drugs affecting potassium channels. Critical for elderly patients and those on multiple QT-prolonging agents.
Hepatotoxicity
Liver damage risk from drug metabolism. Elevated in patients with pre-existing hepatic impairment or on hepatotoxic combinations.
Nephrotoxicity
Kidney damage risk from drug excretion. Higher in elderly, diabetic, or hypertensive patients with reduced renal function.
GI Bleeding
Gastrointestinal bleeding risk from NSAIDs, anticoagulants, and antiplatelet agents. Amplified by polypharmacy.
Serotonin Syndrome
Life-threatening serotonergic excess from drug combinations. Requires immediate recognition and intervention.
Risk Adjustment Factors
| Factor | Multiplier | Clinical Rationale |
|---|---|---|
| Age > 65 years | +2% per year | Reduced organ reserve, altered pharmacokinetics |
| Each comorbidity | +15% | Increased vulnerability, drug-disease interactions |
| Each medication | +10% | Polypharmacy complexity, interaction probability |
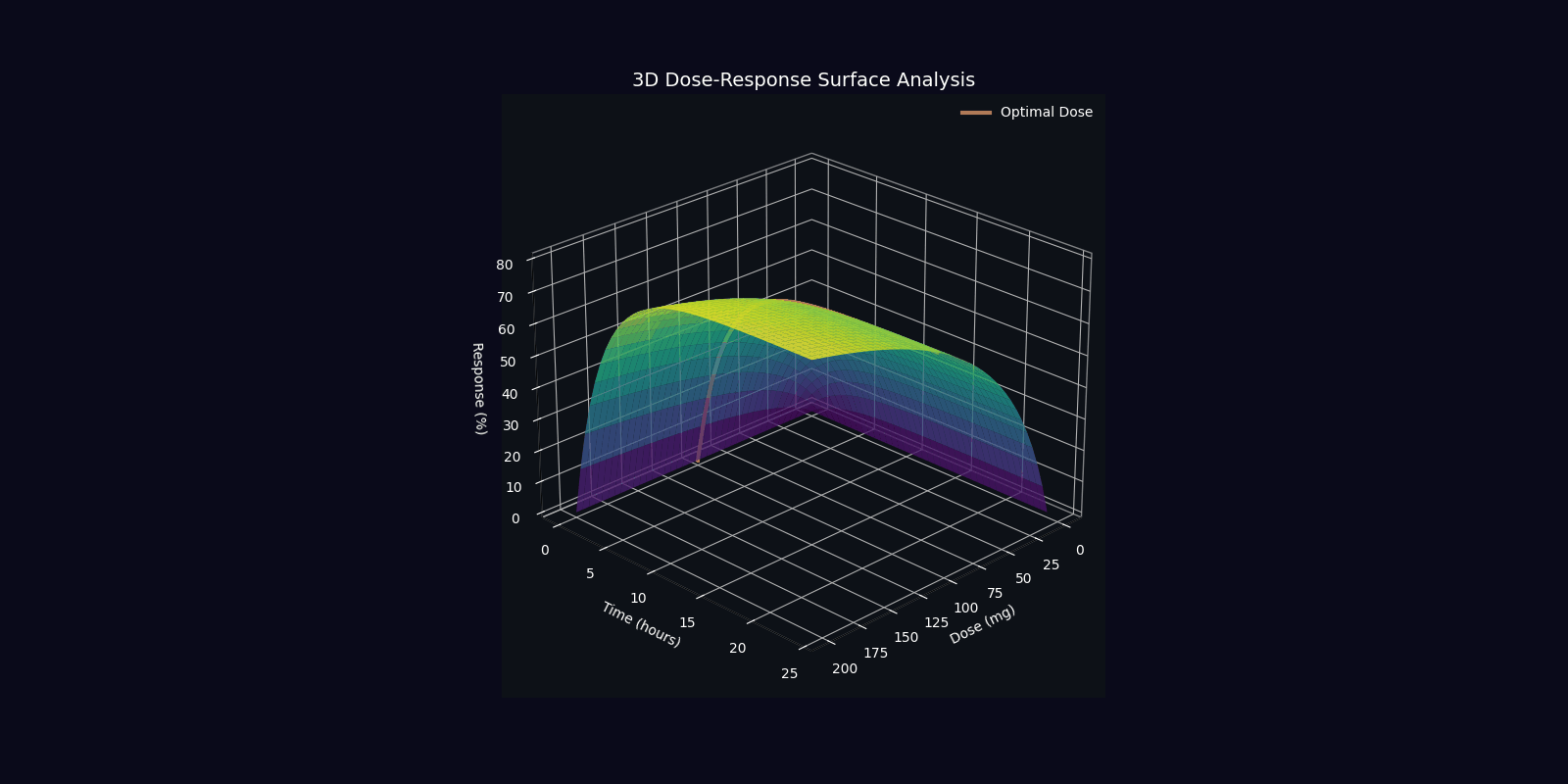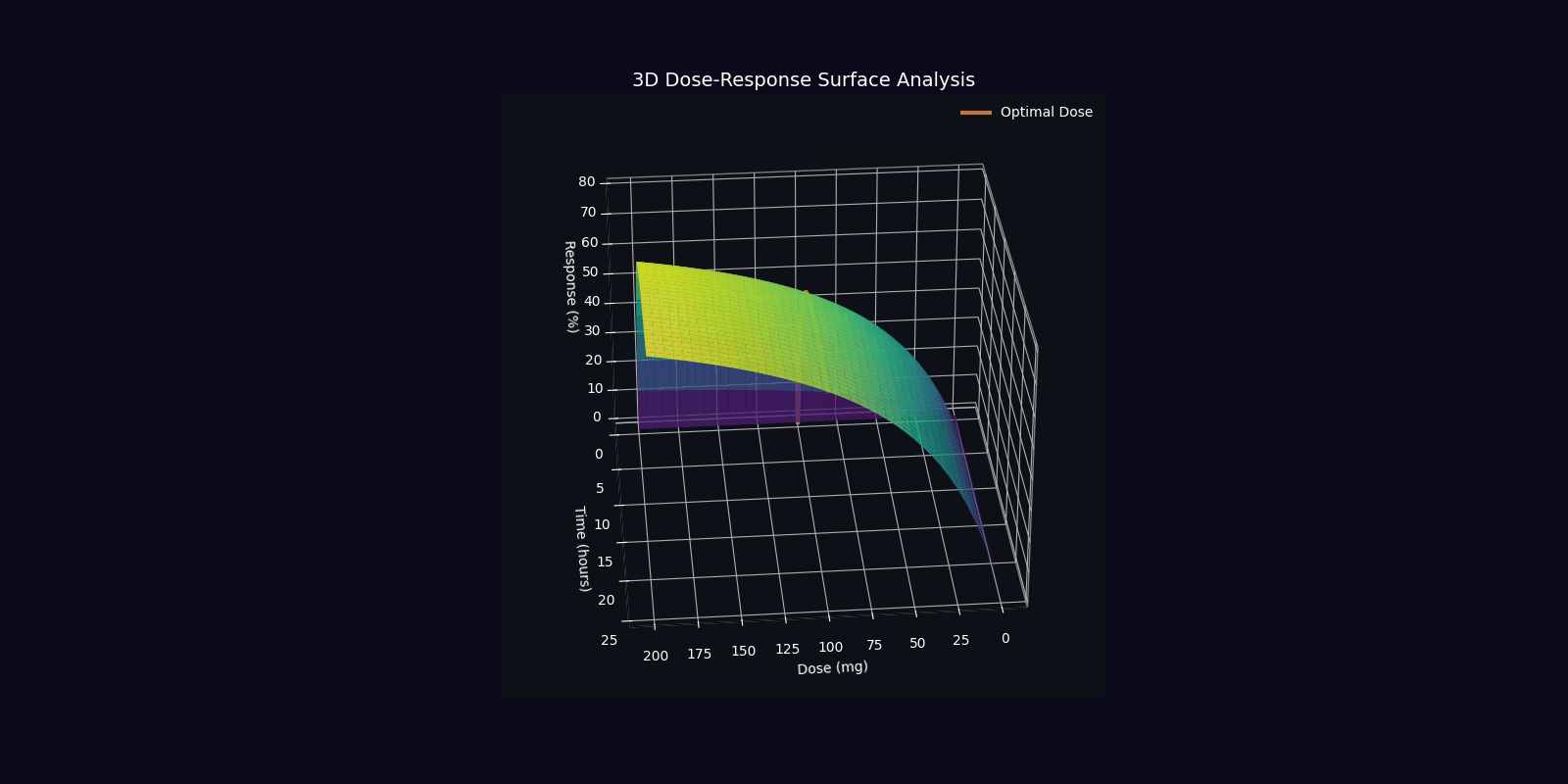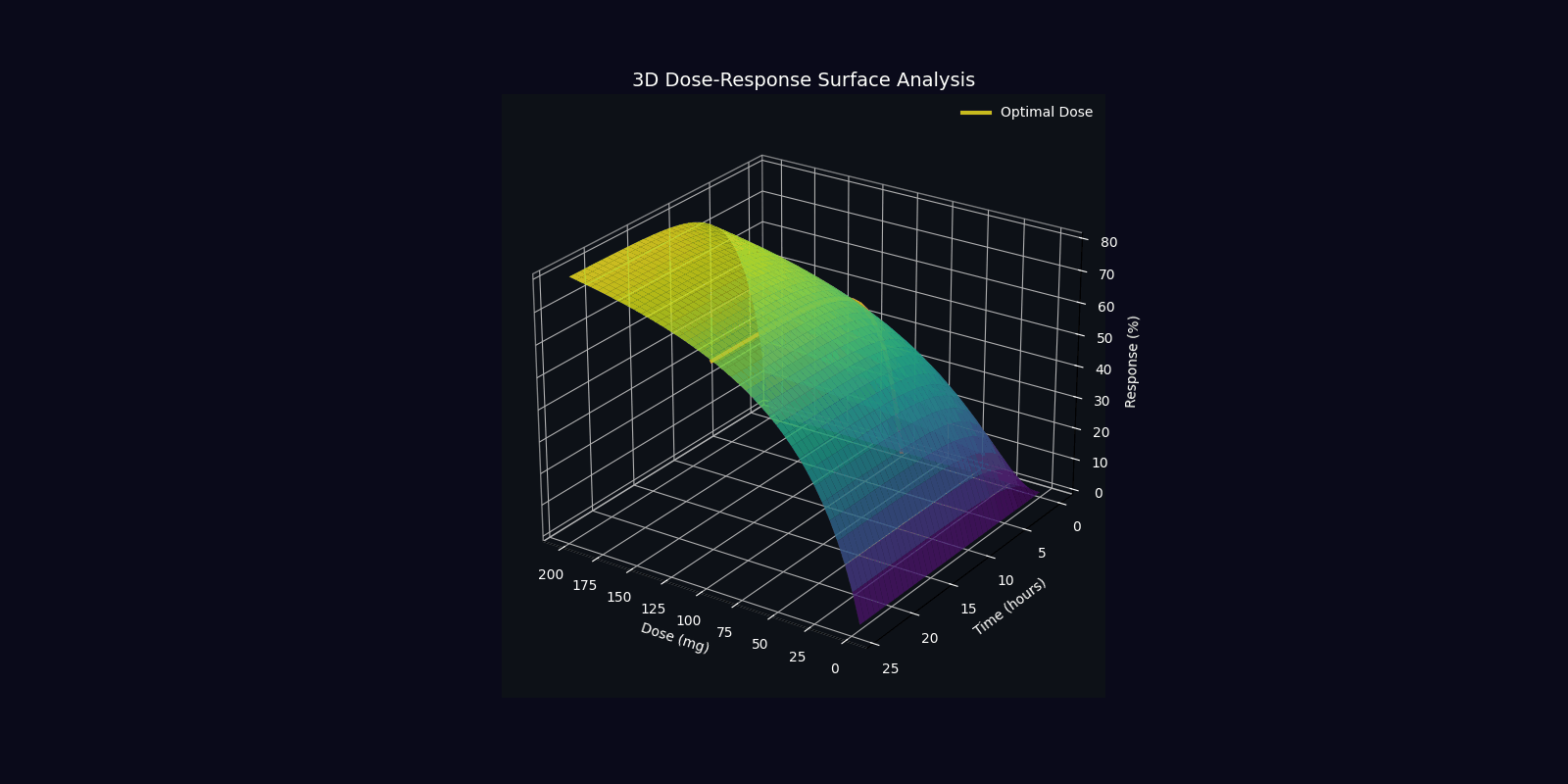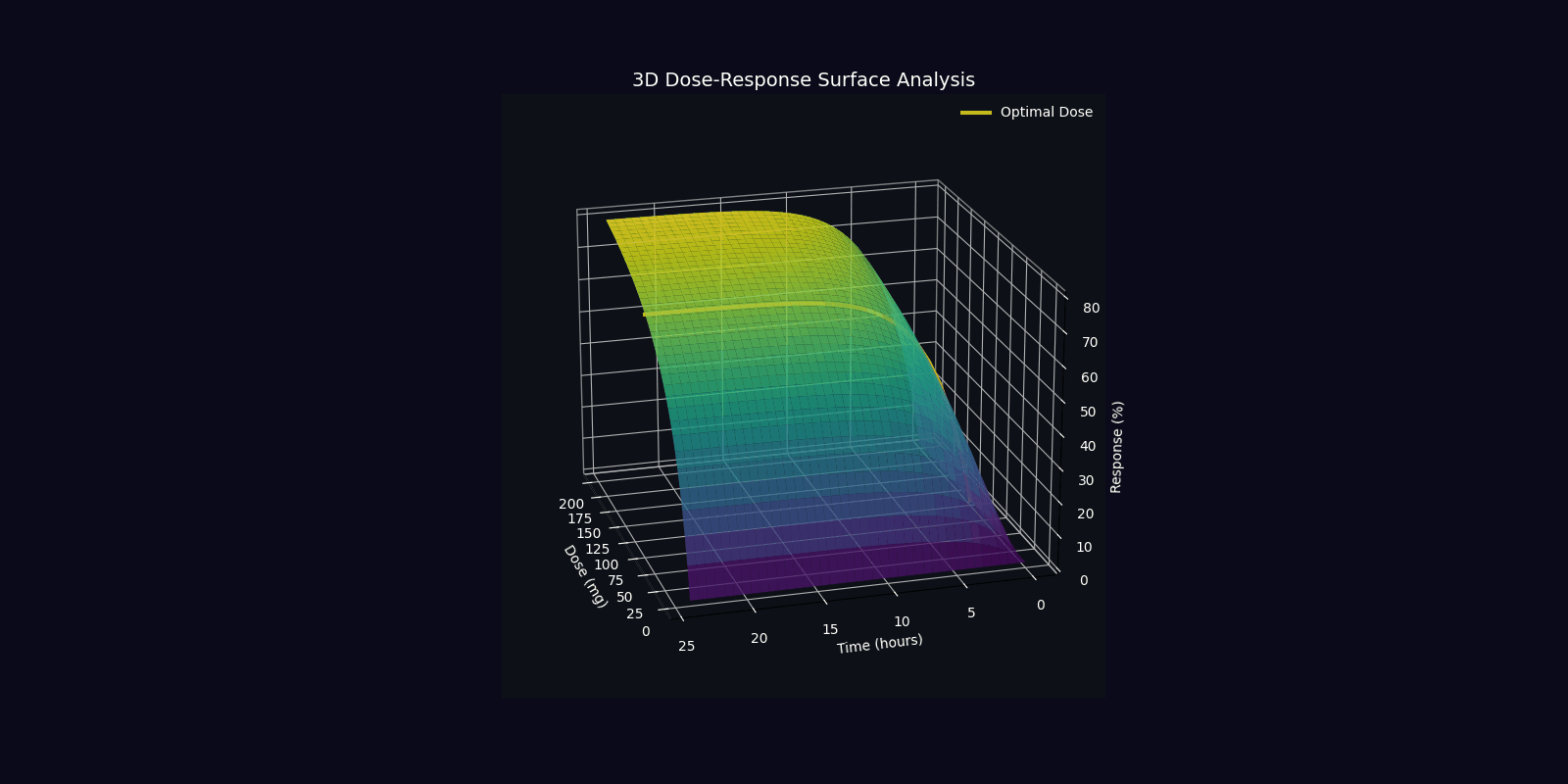
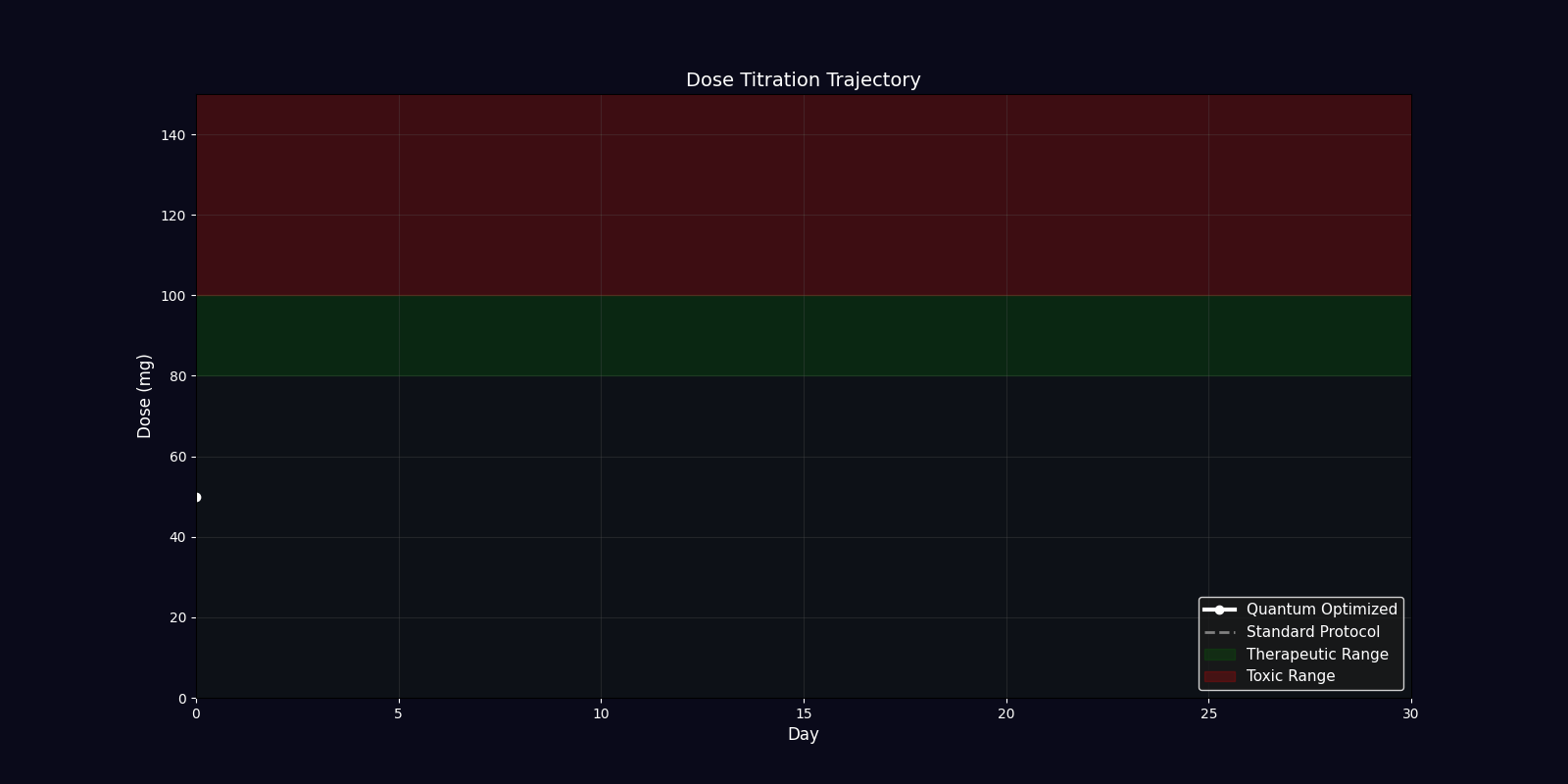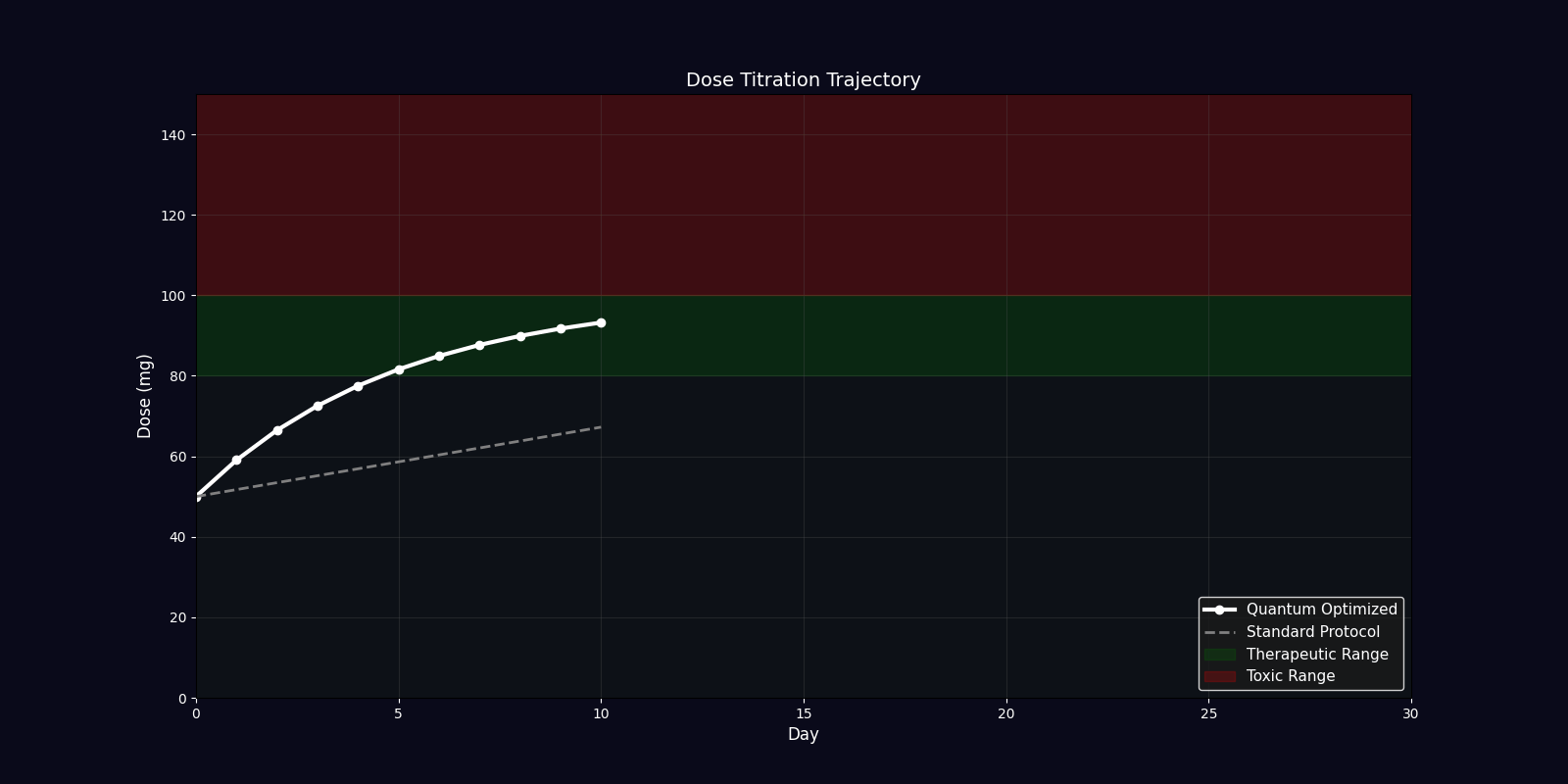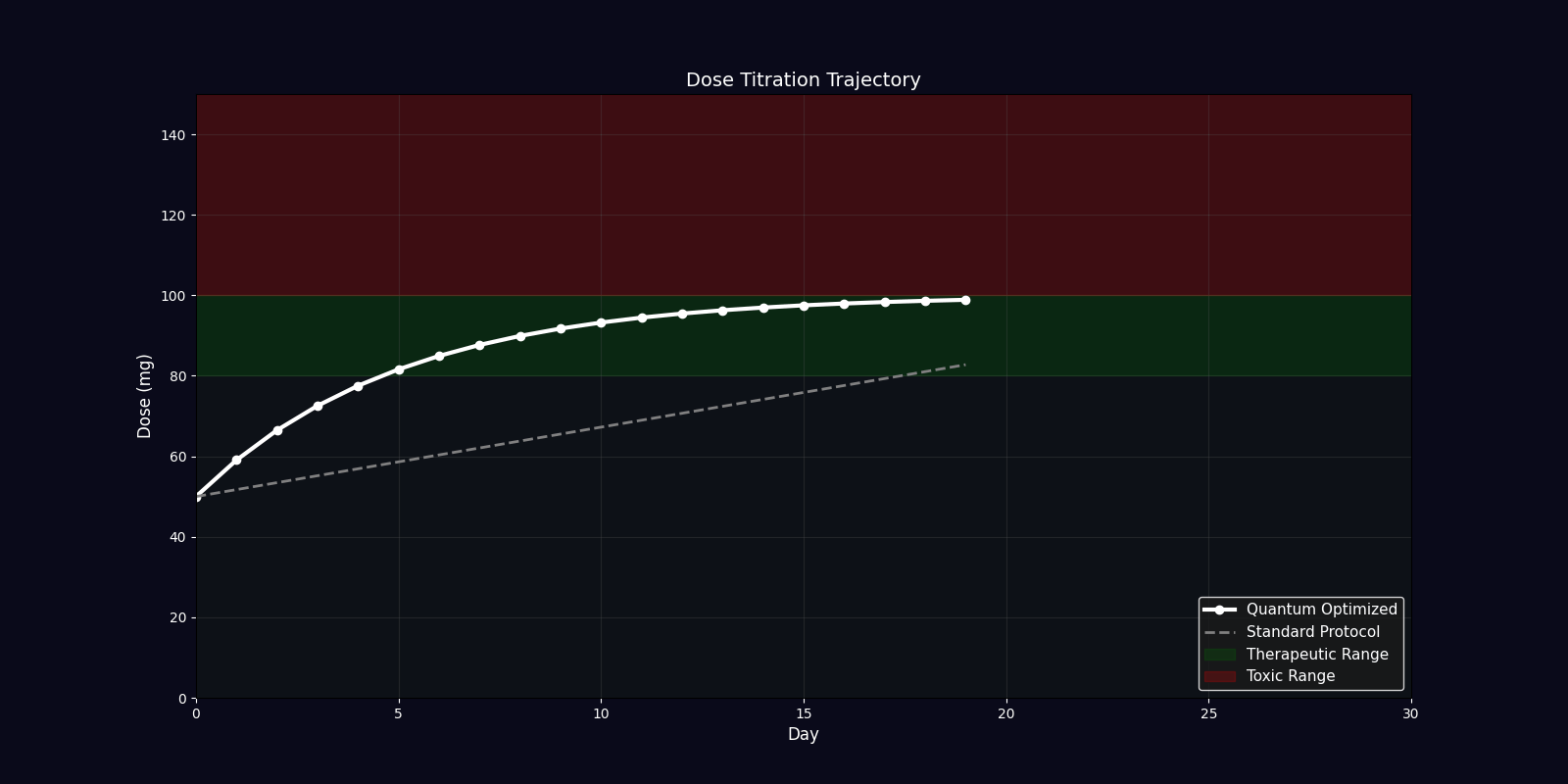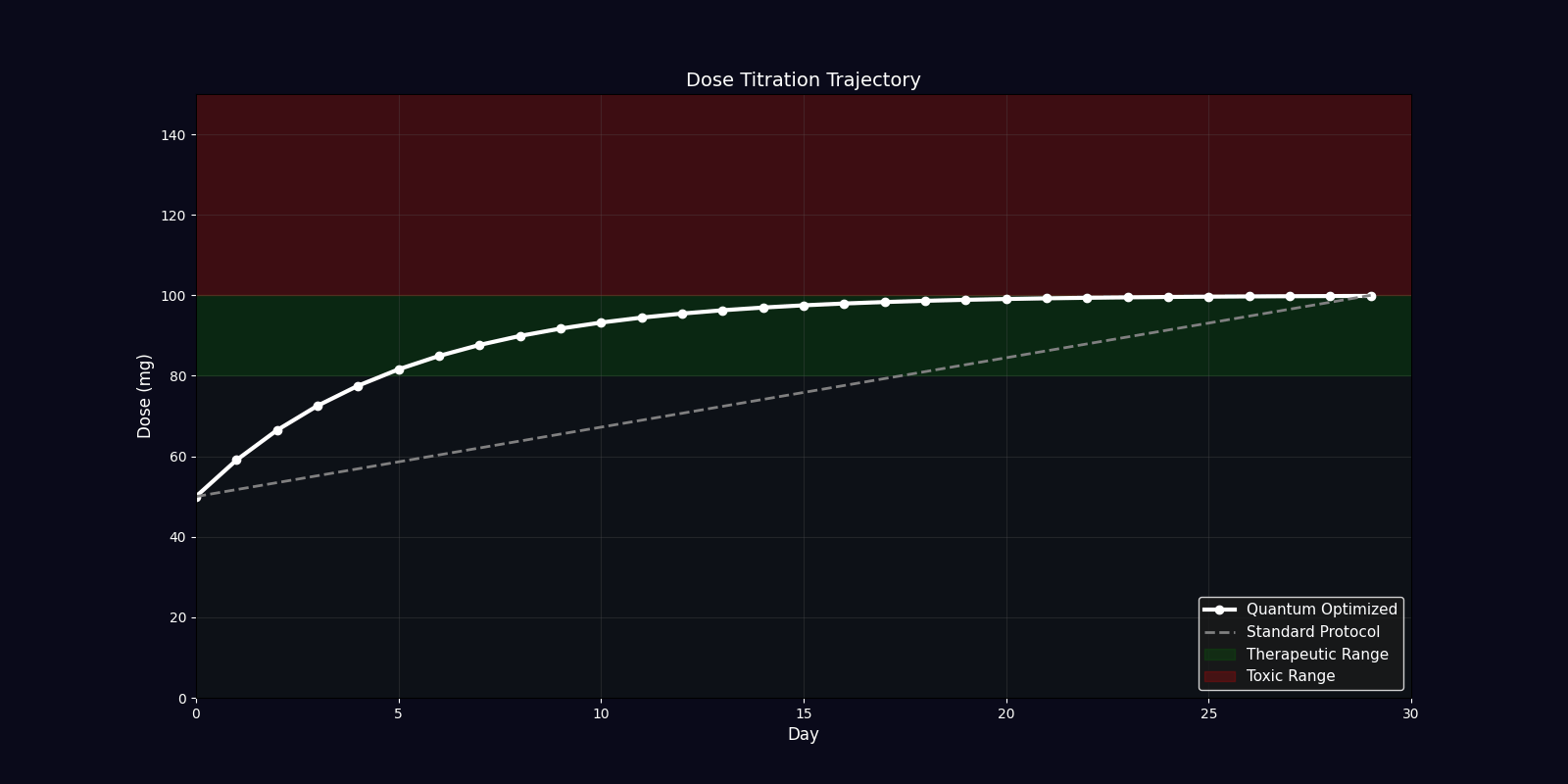
Dynamic Simulations (18 Total: 10 x 3D + 6 x 2D)
QPDP generates 18 comprehensive 4-second GIF simulations (30 FPS, 1920x1080) demonstrating molecular dynamics, quantum states, pharmacokinetics, genomic profiles, and clinical trajectories. Each simulation visualizes complex interactions in personalized medicine.
📁 All 18 simulations available in: QPDP/personalized_medicine_output/ directory
View detailed specifications in Technical Report →
3D Molecular & Protein Simulations (4)
3D Quantum & Clinical Simulations (6)
2D Dynamic Simulations (6)
Technical Analysis Graphs (5 High-Resolution PNG)
Comprehensive static graphs (2100x1500, 150 DPI) providing in-depth analysis of pharmacogenomics, quantum circuits, drug interactions, ADR stratification, and health economics.
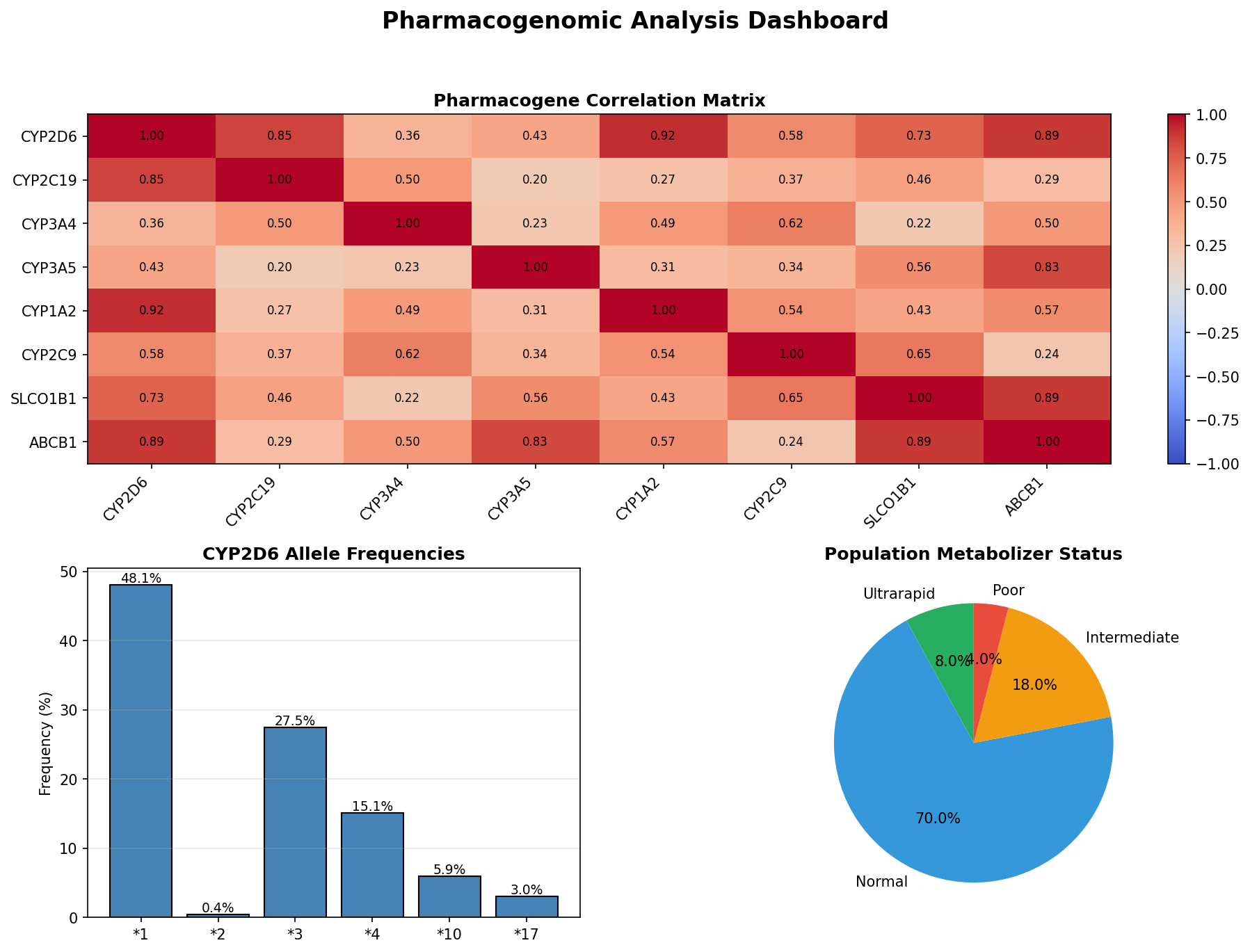
Pharmacogenomic Analysis: 8×8 gene correlation matrix, allele frequencies, metabolizer distribution (800 KB)
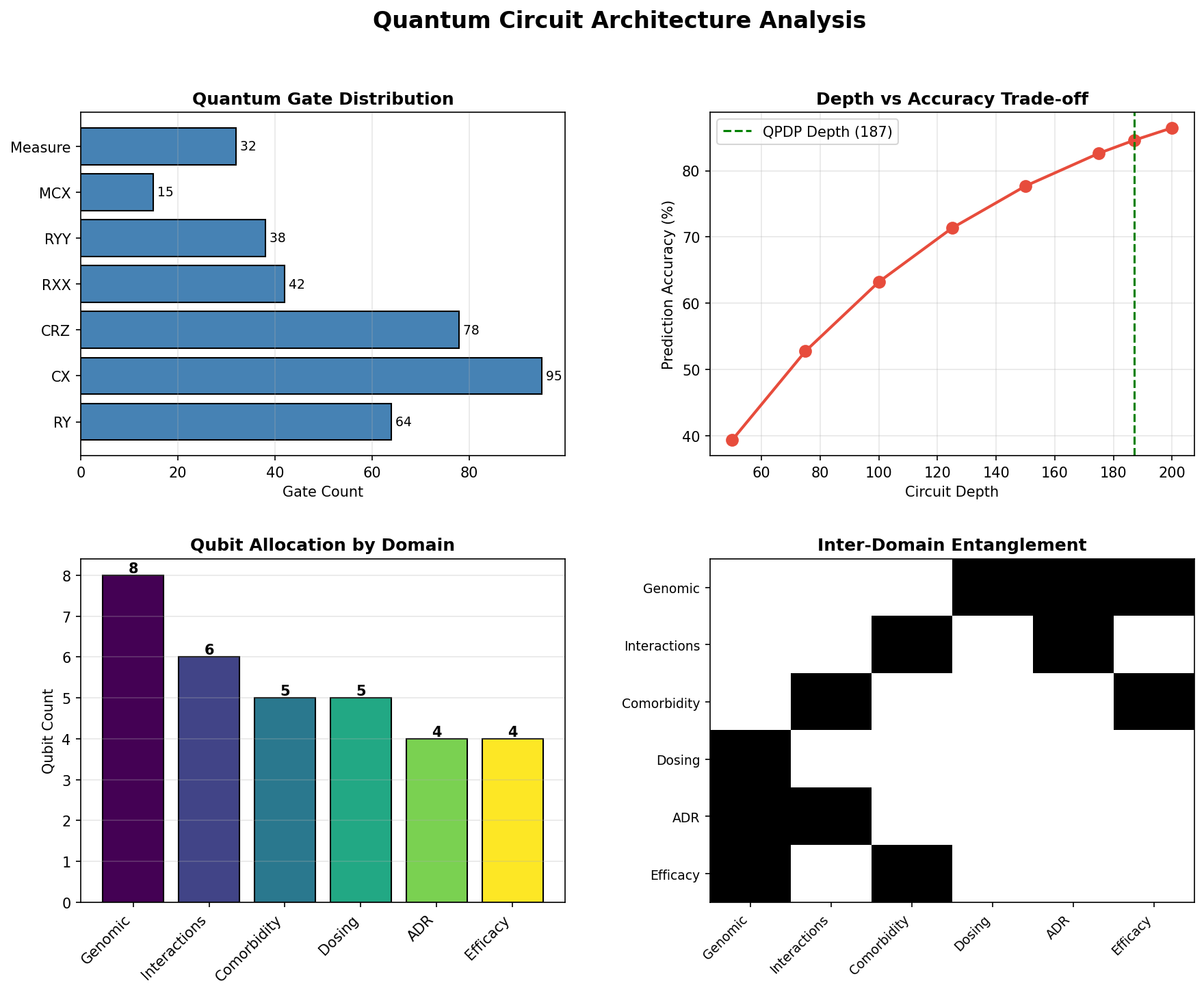
Quantum Circuit Analysis: Gate distribution, depth vs accuracy, qubit allocation, entanglement connectivity (900 KB)
Personalized Medicine Dashboard
Comprehensive 10-panel dashboard integrating patient profile, genomic data, quantum metrics, dosing recommendations, ADR risks, and efficacy predictions.
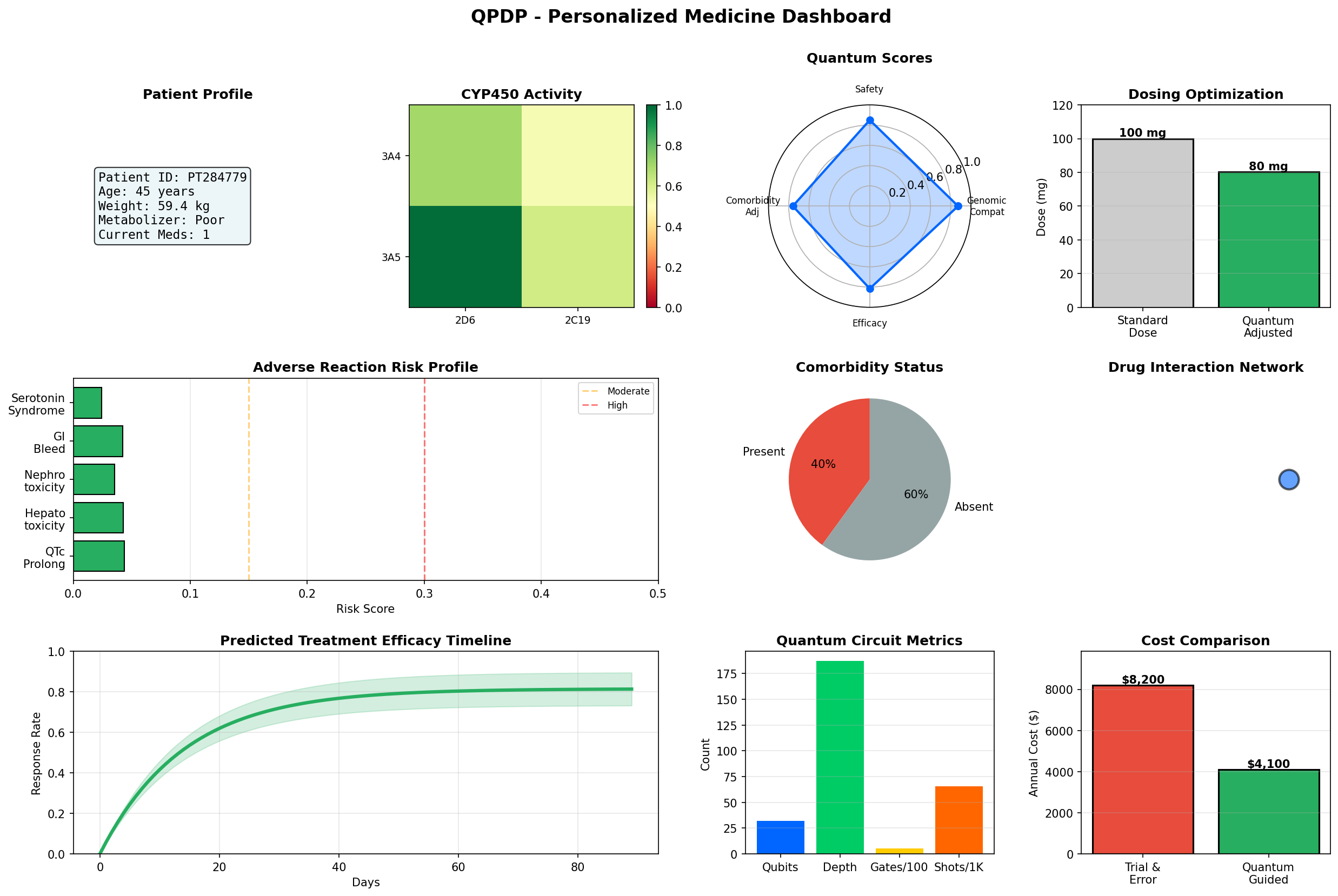
Click to view full dashboard - 10 panels covering patient profile, CYP450 activity, quantum scores, dosing optimization, ADR risks, comorbidity impact, drug interactions, efficacy timeline, circuit metrics, and cost-benefit analysis
Dashboard Components
| Panel | Visualization | Clinical Value |
|---|---|---|
| Patient Profile Summary | Text summary | Quick reference for demographics and metabolizer status |
| CYP450 Activity Heatmap | 2x2 heatmap | Visual enzyme activity levels for 4 key CYP enzymes |
| Quantum Metrics Radar | Polar radar chart | Holistic view of genomic compatibility, safety, and efficacy |
| Dosing Optimization | Bar chart | Standard vs quantum-adjusted dose comparison |
| ADR Risk Breakdown | Horizontal bar chart | Specific ADR risks with color-coded severity |
| Comorbidity Impact | Pie chart | Present vs absent comorbidity distribution |
| Drug Interaction Network | Network graph | Visual representation of polypharmacy interactions |
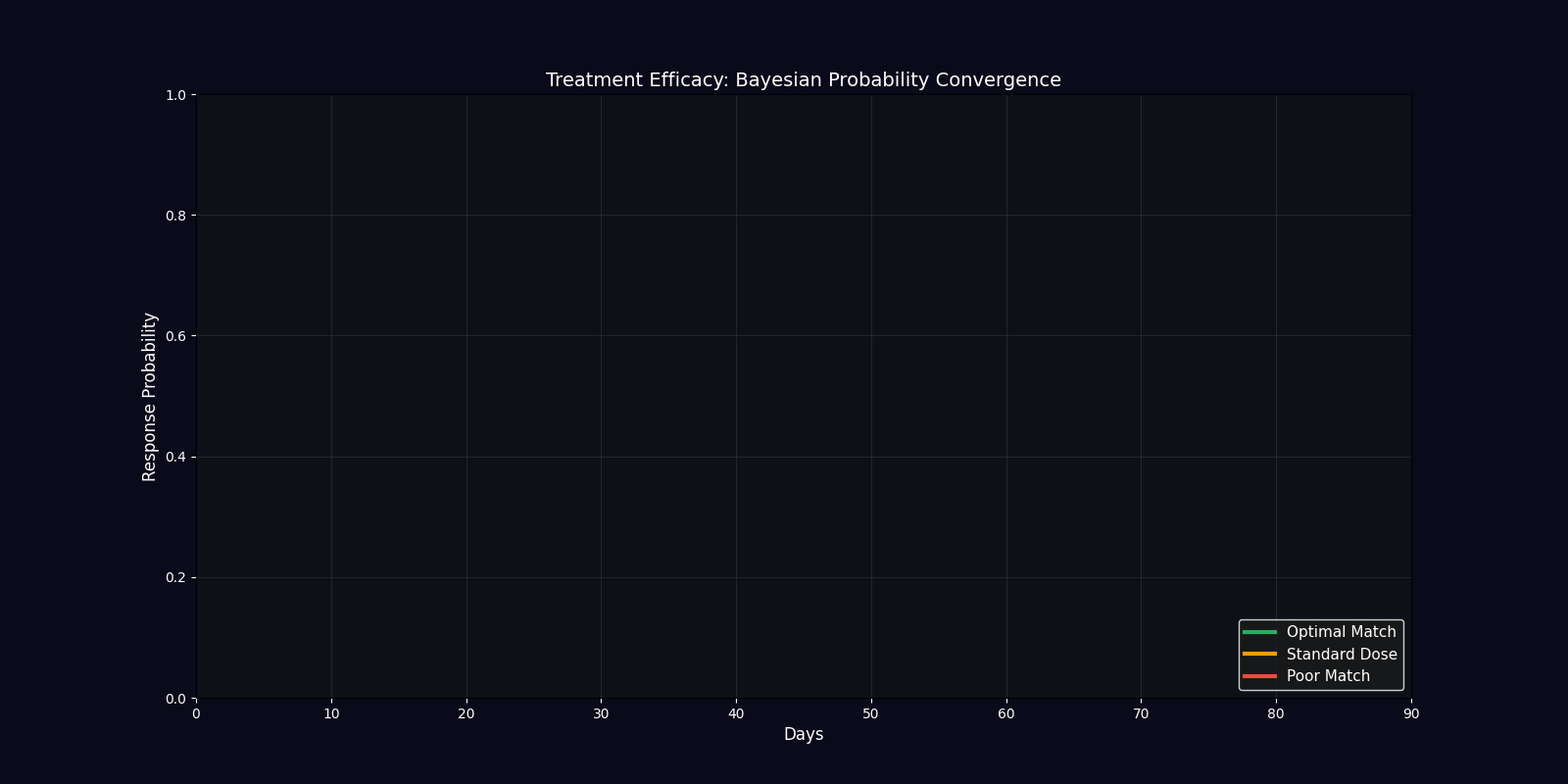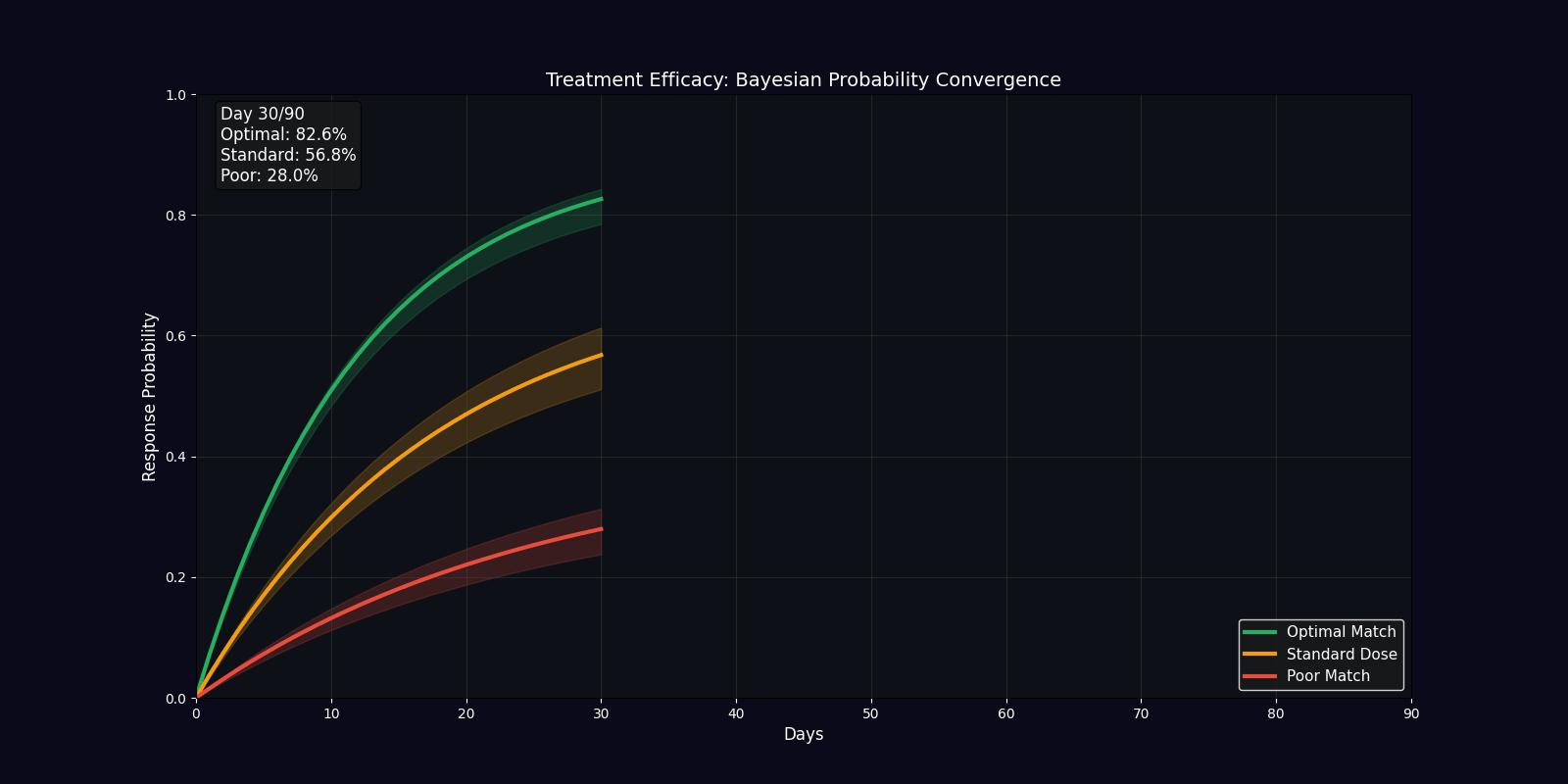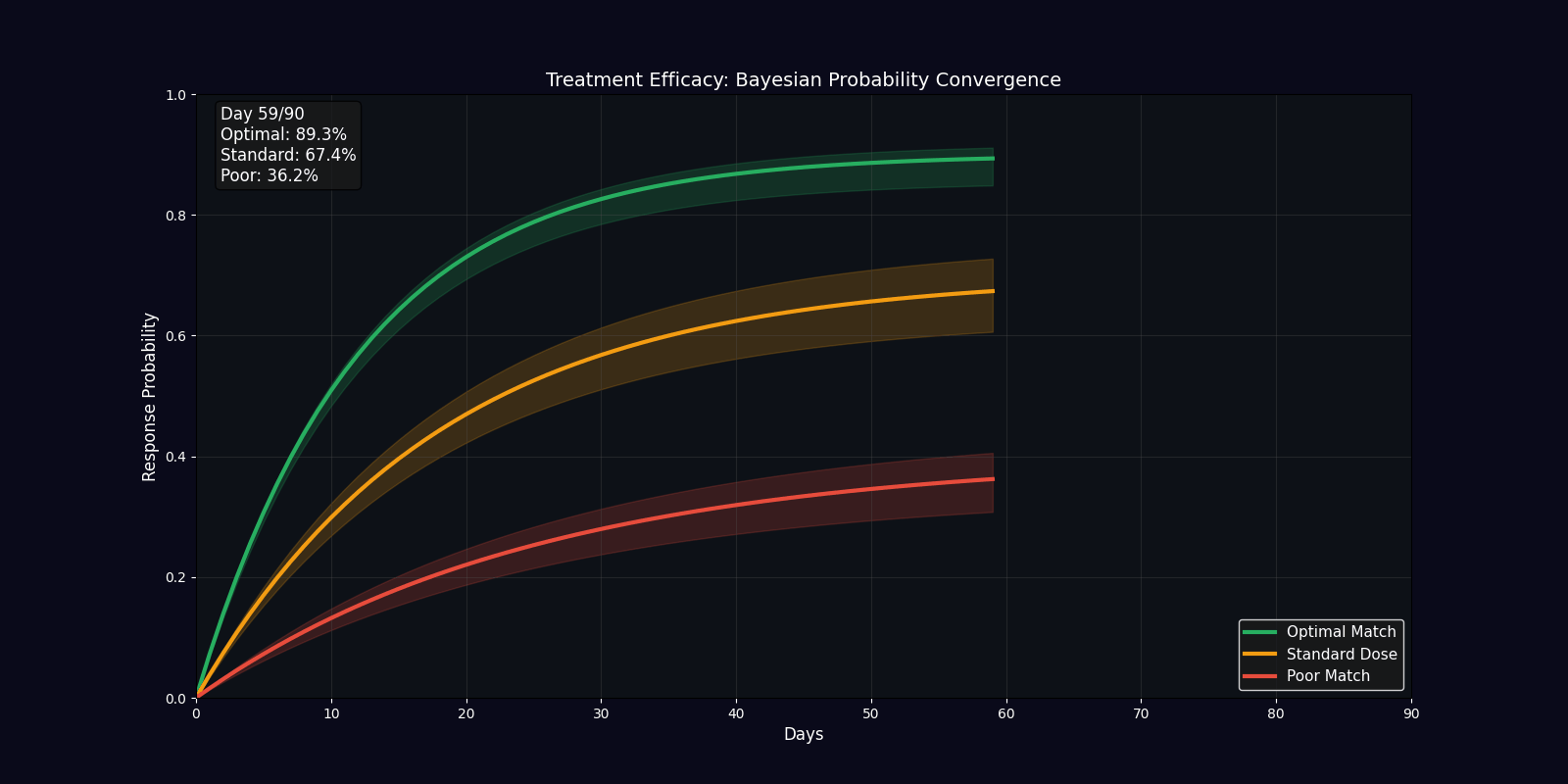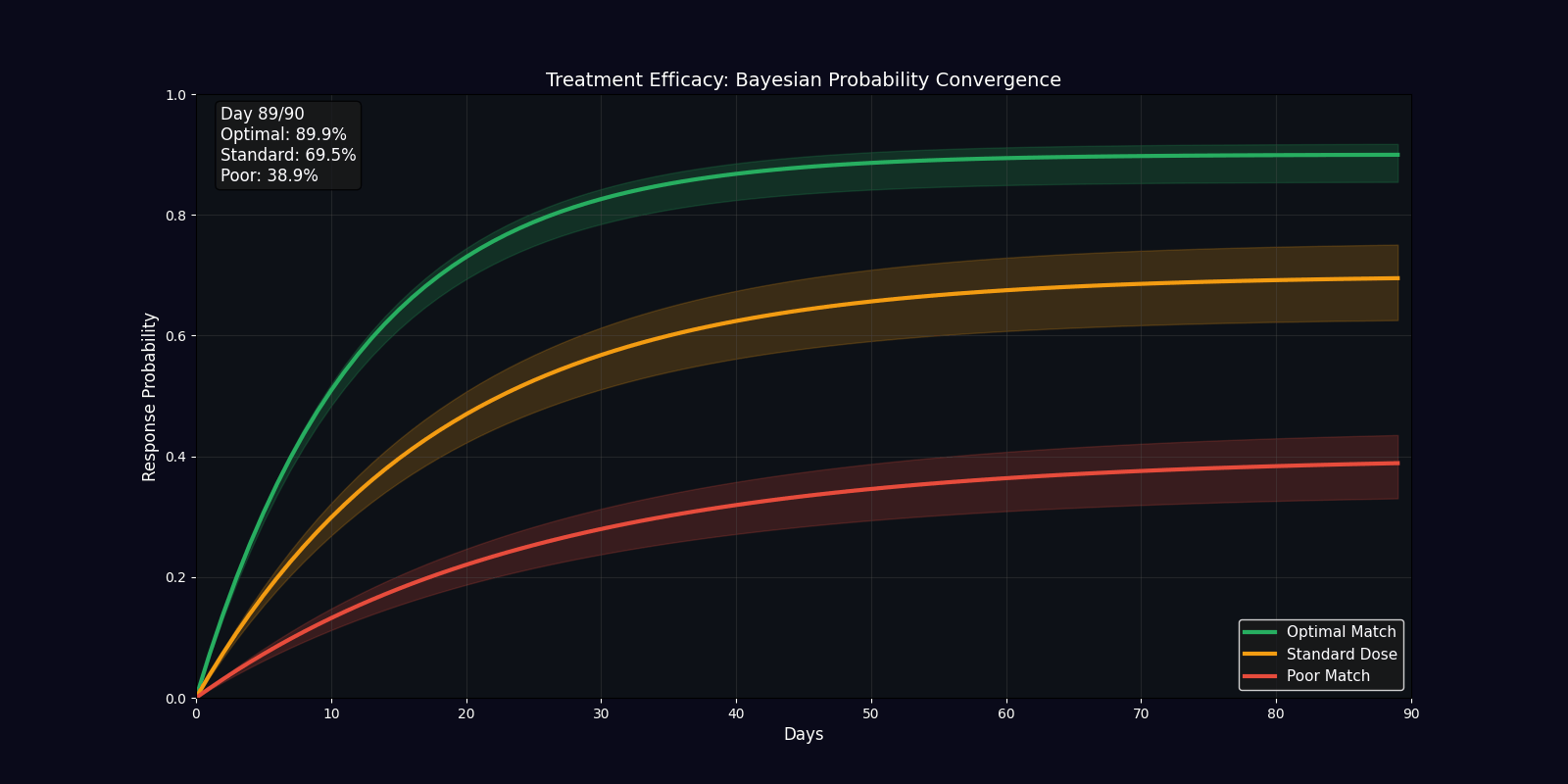
| Efficacy Timeline | Line chart | Predicted treatment response over 90 days |
| Quantum Circuit Stats | Bar chart | Circuit complexity metrics (qubits, depth, gates, shots) |
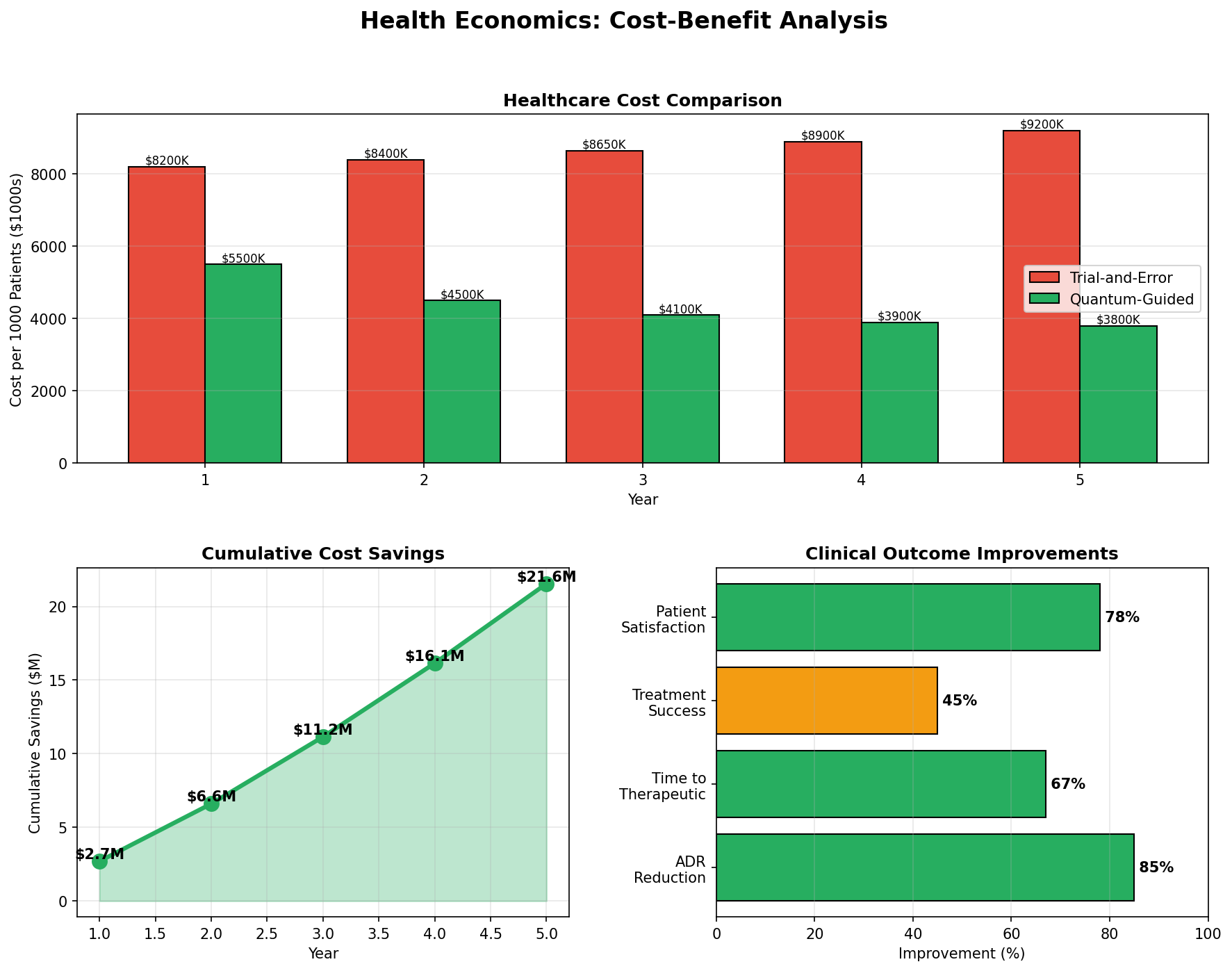
| Cost-Benefit Analysis | Bar chart | Annual healthcare cost comparison: trial-and-error vs quantum-guided |
Output Files
QPDP generates comprehensive output files including PNG dashboards, interactive HTML reports, GIF simulations, JSON data, and text summaries.
Source Code
Complete Python implementation with all accumulated features from QFRM, QFDNA, QPPIP, and QSCLO plus healthcare-specific capabilities.
qpdp.py
Main quantum personalized medicine engine with 32-qubit circuit, genomic profiling, drug interaction network, ADR risk calculator, and visualization engine.
View on GitHubKey Libraries & Technologies
| Category | Libraries | Purpose |
|---|---|---|
| Quantum Computing | Qiskit, Qiskit-Aer, Qiskit-Algorithms | 32-qubit circuit design and simulation |
| Data Science | NumPy, Pandas, SciPy, scikit-learn | Data processing, statistical analysis |
| Visualization | Matplotlib, Plotly, Seaborn | Dashboards, interactive reports, simulations |
| Graph Analysis | NetworkX | Drug interaction network modeling |
| Performance | psutil, Pillow | System monitoring, GIF generation |